Difference between revisions of "CNF Imaging Workshop"
(→Schedule) |
(→Registration) |
||
| (54 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
= Overview = | = Overview = | ||
| − | |||
| − | |||
| − | Next generation Imaging for Nuclear Femtography is in its very early stages. The complexity from seven-dimensional data and many scales and levels of interactions between the colliding particles and what is observed create many challenges. To address these challenges the “Next-generation imaging filters and mesh-based data representation for phase-space calculations in nuclear femtography (CNF19-04)” proposed to put together an interdisciplinary team to: | + | [[File:Cnf-workshop-logo-with-letters.png||right|255px]] |
| + | |||
| + | Next generation Imaging for Nuclear Femtography is in its very early stages. The complexity from seven-dimensional data and many scales and levels of interactions between the colliding particles and what is observed create many challenges. To address these challenges the “Next-generation imaging filters and mesh-based data representation for phase-space calculations in nuclear femtography (CNF19-04)” project proposed to put together an interdisciplinary team to: | ||
| + | * leverage advanced computational methods and software design approaches from medical imaging for processing phase space distributions in nuclear femtography experiments, with the long term aim to enable next-generation process simulations, data analyses, and physics model comparisons and | ||
| + | * organize a mini-workshop that will: (a) bring world-wide leaders from Imaging in Health Care and (b) nation- and state-wide experts in transferring technology to industry in VA. | ||
| − | Two of the most advanced groups world-wide in Medical Image Computing | + | Two of the most advanced groups world-wide in Medical Image Computing and Image Guided Neurosurgery are in Brigham and Women's Hospital (BWH), Harvard Medical School. Namely the Surgical Planning Laboratory (SPL) lead by Dr. Ron Kikinis and Image Guided Neurosurgery (IGNS) Laboratory lead by Dr. Alexandra Golby. |
| − | |||
| − | + | *'''Monday, August 19,''' the objective is to focus on '''Lessons Learned from Medical Image Computing''' at SPL and IGNS at Harvard; together both labs have more than 40 years of experience and many 100’s of person-years of accumulated research. | |
| + | Job creation and transfer of technology to industry is as crucial for sustaining basic research as the quality of research in general and specifically in Nuclear Femtography. However, nurturing and flexibility as it is demonstrated at the Institute for Advanced Study at Princeton and argued in the “Usefulness of Useless Knowledge”[https://library.ias.edu/files/UsefulnessHarpers.pdf] is required. | ||
| + | *'''Tuesday, August 20''' the objective is to focus on '''Lessons Learned form empirical evidence''', address economic strengths and weaknesses of various approaches to generating inventions and innovations, in part address intellectual property and protection --a key part of the process for job creation-- and focus on VA ecosystem, state-based incentives and programs for innovation. Tuesday’s discussion will be lead by Prof. James Koch. Mr. James Cheng and Dr. Steve Pieper. | ||
| − | + | The CNF Imaging Workshop expected to be '''highly interactive''' as participants will transfer know-how from the medical imaging community to basic physics in this case nuclear femtography. | |
| − | |||
{| style="border-spacing: 2px;" | {| style="border-spacing: 2px;" | ||
! style="width: 400px;" | | ! style="width: 400px;" | | ||
| Line 26: | Line 28: | ||
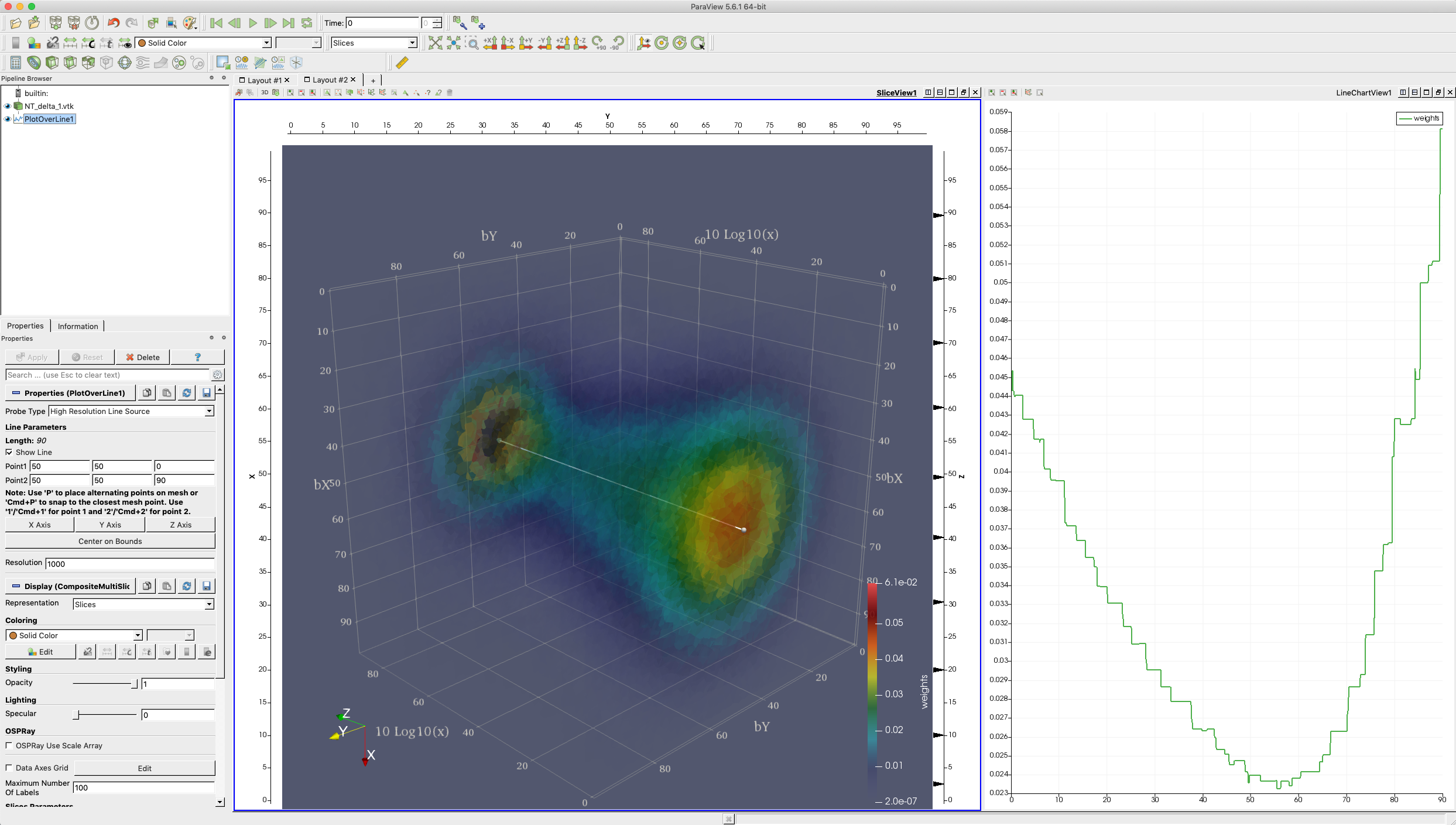
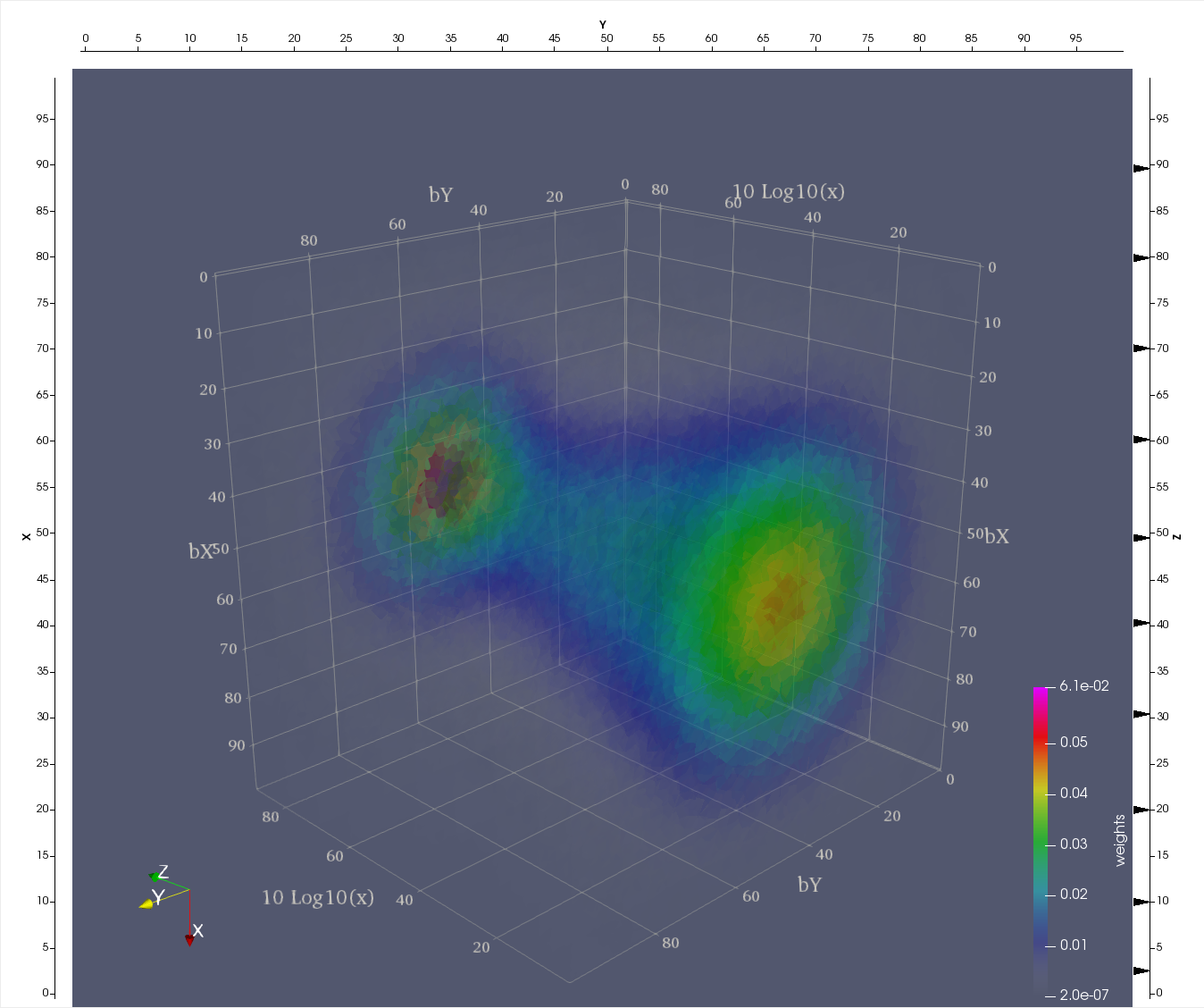
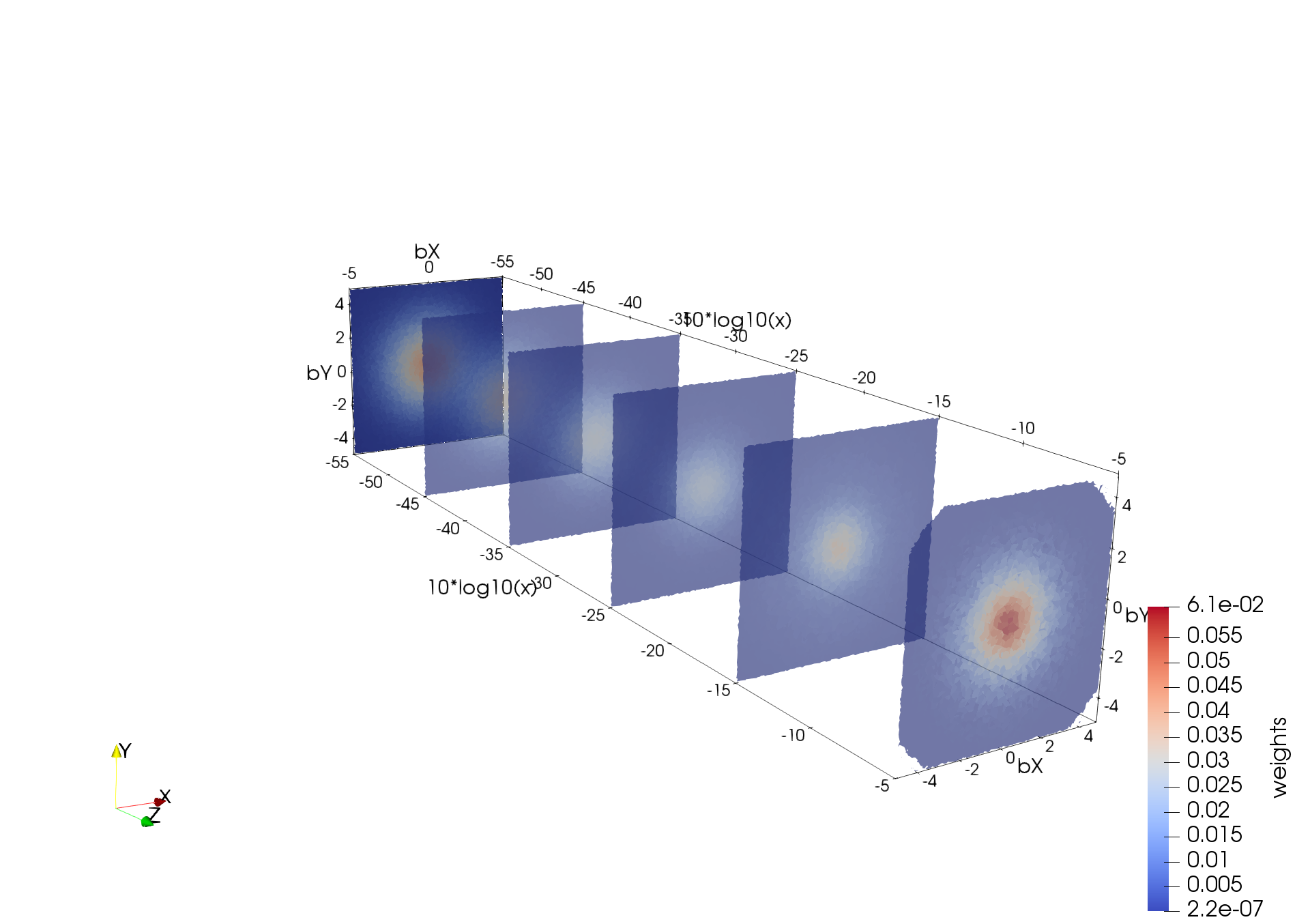
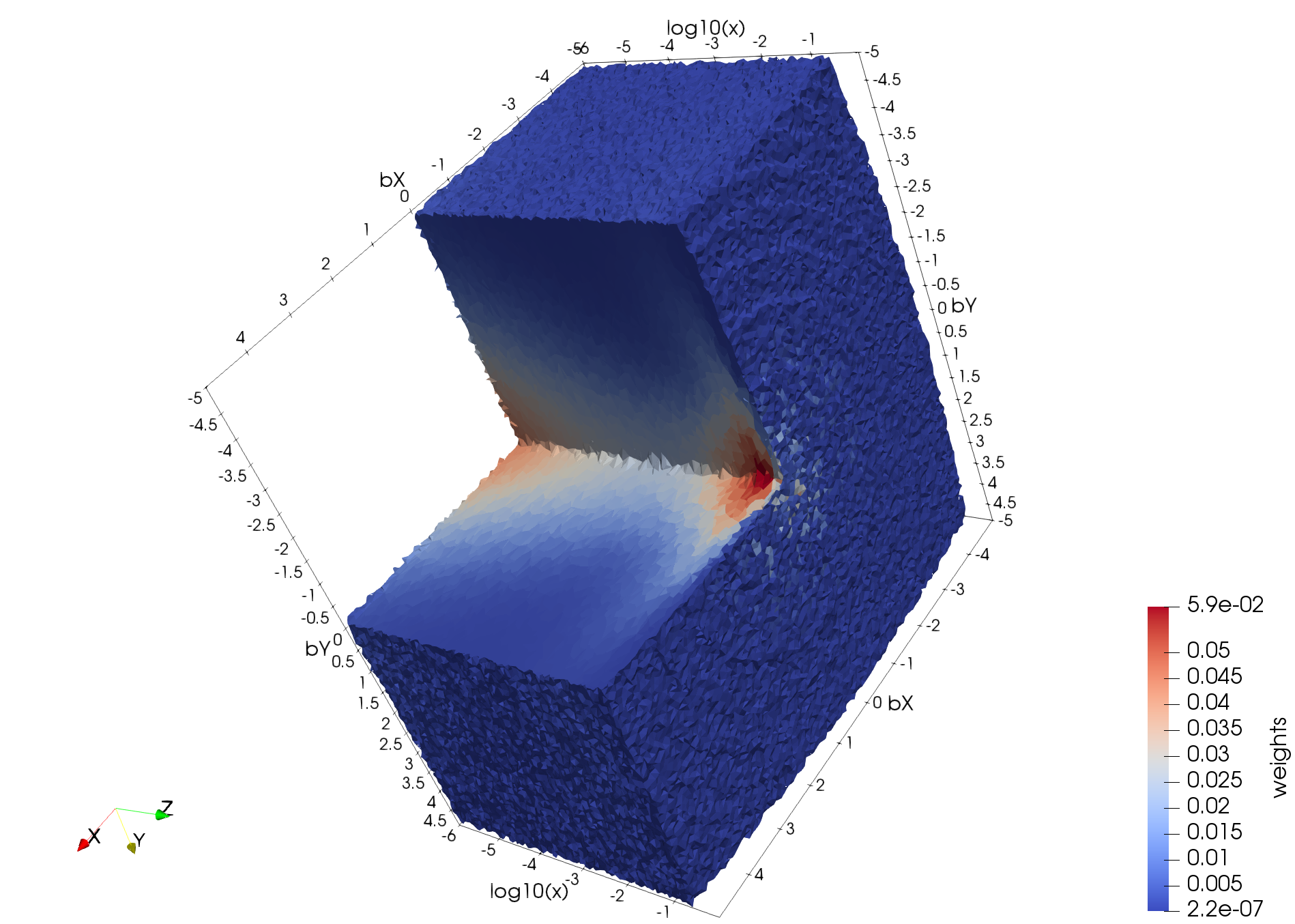
The tessellation data in figure (above) depict a spatial distribution of up quarks as a function of proton's momentum fraction carried by those quarks; | The tessellation data in figure (above) depict a spatial distribution of up quarks as a function of proton's momentum fraction carried by those quarks; | ||
| − | bX and bY, spatial coordinates (in 1/GeV = 0.197 fm) defined in a plane perpendicular to the nucleon’s motion, x is the fraction of proton’s momentum and color denotes probability density for finding a quark at given (bX, bY, x). These preliminary data are generated by Dr. Sznajder and processed/tessellated with CRTC's CNF_I2M tool. Their visualization is accomplished by Dr. Gavalian using Paraview. | + | bX and bY, spatial coordinates (in 1/GeV = 0.197 fm) defined in a plane perpendicular to the nucleon’s motion, x is the fraction of proton’s momentum and color denotes probability density for finding a quark at given (bX, bY, x). These preliminary data are generated by Dr. Sznajder and processed/tessellated with CRTC's CNF_I2M tool. Their visualization is accomplished by Dr. Gavalian using Paraview. |
| + | = Schedule = | ||
| + | '''August 19:''' (Discuss lessons learned from the medical imaging discipline) | ||
| + | |||
| + | * Morning Session: 9:30AM to 12:30 (Chair: Christian) | ||
| + | ** Welcome and Logistics for the workshop (Nikos): 9:30AM to 9:40AM | ||
| + | ** Introduction to Jefferson Lab (Bob): 9:40AM to 9:55AM | ||
| + | ** Brief review of CoS at ODU's efforts with CNF Initiative (Gail): 9:55AM to 10:10AM | ||
| + | ** Introduction to Center for Nuclear Femtography (David): 10:10AM to 10:25AM | ||
| + | ** Imaging subnuclear systems: Concepts, objectives, applications (Christian): 10:25AM to 10:45AM | ||
| + | ** '''External Speakers:''' Ron Kikinis and Alexandra Golby: 10:45AM to 12:30PM | ||
| + | |||
| + | * Lunch Break: 12:30PM to 1:30PM | ||
| − | + | * Afternoon Session: 1:30PM to 3:45PM (Chair: Gagik) | |
| − | + | ** Focus on software design and building an ecosystem and community around 3D Slicer | |
| + | ** '''External Speakers:'''Tina Kapur, Steve Pieper, and Andrey Fedorov | ||
| + | |||
| + | * Dinner at 7:30PM -- TBD | ||
| + | |||
| + | '''August 20:''' Broader Impact to the Community | ||
| + | |||
| + | * Morning BoF Session : 10:30AM to 12:00PM (Chair: Nikos) | ||
| + | ** Focus on Empirical Data and Lessons Learned on Innovation broadly and more specifically in VA. | ||
| + | ** '''External Speakers:''' James Koch, James Cheng and Steve Pieper | ||
| + | |||
| + | * Lunch Break: 12:00 to 1:30PM | ||
| + | |||
| + | * Afternoon Session: 1:30PM to 3:30PM (Chair: Christos/Nikos) | ||
| + | ** Focus on JLAB projects with concrete examples on imaging CNF data | ||
| + | *** Experimental Physicist's Point of View on Imaging for Nuclear Femtography (Gagik): 1:30PM to 2:00PM | ||
| + | *** Discussion Moving Forward (Open to all attendees and to be moderated by Nikos): 2:00PM to 3:30PM | ||
| + | |||
| + | = Registration = | ||
| + | '''Register here:''' https://cnf_imaging_workshop.eventbrite.com | ||
= Presenters = | = Presenters = | ||
| Line 38: | Line 71: | ||
<li style="display: inline-block;"> | <li style="display: inline-block;"> | ||
[[File: Ron.png|thumb|left|500px| '''Dr. Ron Kikinis: Center Director, Neuroimaging Analysis Center, Director, Surgical Planning Laboratory, Professor of Radiology at Harvard Medical School''']] | [[File: Ron.png|thumb|left|500px| '''Dr. Ron Kikinis: Center Director, Neuroimaging Analysis Center, Director, Surgical Planning Laboratory, Professor of Radiology at Harvard Medical School''']] | ||
| − | '''Dr. Ron Kikinis is the Robert Greenes Distinguished Director of Biomedical Informatics in the Department of Radiology at at Harvard Medical School and "Institutsleiter" of Fraunhofer MEVIS and Honorary Professor of Medical Image Computing at the University of Bremen.''' He is the founding Director of the Surgical Planning Laboratory, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, and a Professor of Radiology at Harvard Medical School. This laboratory was founded in 1990. Before joining Brigham & Women's Hospital in 1988, he trained as a resident in radiology at the University Hospital in Zurich, and as a researcher in computer vision at the ETH in Zurich, Switzerland. He received his M.D. degree from the University of Zurich, Switzerland, in 1982. In 2004 he was appointed Professor of Radiology at Harvard Medical School. In 2009 he was the inaugural recipient of the MICCAI Society "Enduring Impact Award". On February 24, 2010 he was appointed the Robert Greenes Distinguished Director of Biomedical Informatics in the Department of Radiology at Brigham and Women's Hospital. On January 1, 2014, he was appointed "Institutsleiter" of Fraunhofer MEVIS and Professor of Medical Image Computing at the University of Bremen. Since then he is commuting every two months between Bremen and Boston. | + | '''Dr. Ron Kikinis is the Robert Greenes Distinguished Director of Biomedical Informatics in the Department of Radiology at at Harvard Medical School and "Institutsleiter" of Fraunhofer MEVIS and Honorary Professor of Medical Image Computing at the University of Bremen.''' '''He is the founding Director of the Surgical Planning Laboratory''', Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, and a Professor of Radiology at Harvard Medical School. This laboratory was founded in 1990. Before joining Brigham & Women's Hospital in 1988, he trained as a resident in radiology at the University Hospital in Zurich, and as a researcher in computer vision at the ETH in Zurich, Switzerland. He received his M.D. degree from the University of Zurich, Switzerland, in 1982. In 2004 he was appointed Professor of Radiology at Harvard Medical School. In 2009 he was the inaugural recipient of the MICCAI Society "Enduring Impact Award". On February 24, 2010 he was appointed the Robert Greenes Distinguished Director of Biomedical Informatics in the Department of Radiology at Brigham and Women's Hospital. On January 1, 2014, he was appointed "Institutsleiter" of Fraunhofer MEVIS and Professor of Medical Image Computing at the University of Bremen. Since then he is commuting every two months between Bremen and Boston. |
During the mid-80's, Dr. Kikinis developed a scientific interest in image processing algorithms and their use for extracting relevant information from medical imaging data. Since then, this topic has matured from a fairly exotic topic to a field of science. This is due to the explosive increase of both the quantity and complexity of imaging data. Dr. Kikinis has led and has participated in research in different areas of science. His activities include technological research (segmentation, registration, visualization, high performance computing), software system development, and biomedical research in a variety of biomedical specialties. The majority of his research is interdisciplinary in nature and is conducted by multidisciplinary teams. The results of his research have been reported in a variety of peer-reviewed journal articles. He is an author and co-author of over 300 peer-reviewed articles. | During the mid-80's, Dr. Kikinis developed a scientific interest in image processing algorithms and their use for extracting relevant information from medical imaging data. Since then, this topic has matured from a fairly exotic topic to a field of science. This is due to the explosive increase of both the quantity and complexity of imaging data. Dr. Kikinis has led and has participated in research in different areas of science. His activities include technological research (segmentation, registration, visualization, high performance computing), software system development, and biomedical research in a variety of biomedical specialties. The majority of his research is interdisciplinary in nature and is conducted by multidisciplinary teams. The results of his research have been reported in a variety of peer-reviewed journal articles. He is an author and co-author of over 300 peer-reviewed articles. | ||
As part of his service to the community, he serves as reviewer for a large number of journals and conferences from the fields of imaging, biomedical engineering and, computer science. He has served and is serving as member of external advisory boards for a variety of centers and research efforts. He is the Principal Investigator of 3D Slicer, a free open source software platform for image analysis and visualization. | As part of his service to the community, he serves as reviewer for a large number of journals and conferences from the fields of imaging, biomedical engineering and, computer science. He has served and is serving as member of external advisory boards for a variety of centers and research efforts. He is the Principal Investigator of 3D Slicer, a free open source software platform for image analysis and visualization. | ||
| Line 111: | Line 144: | ||
=== David Richards === | === David Richards === | ||
<li style="display: inline-block;"> | <li style="display: inline-block;"> | ||
| − | [[File: david_r.jpg|thumb|left|650px| '''David Richards: | + | [[File: david_r.jpg|thumb|left|650px| '''David Richards: Theoretical and Computational Physics at DOE's Jefferson Lab.''']] |
| − | '''Dr. David Richards is | + | '''Dr. David Richards is Theoretical and Computational Physics at DOE's Jefferson Lab.''' Richards came to Jefferson Lab as a staff scientist and joint faculty member at Old Dominion University in 1999. He became a full-time staff scientist in 2002 and served as acting Theory Center leader from September 2009 through October 2010. He was appointed deputy director of the Theory Center in mid-October 2010. Richards' current research focus is aimed at garnering a better understanding of so-called "excited states." These are subatomic particles that were once the familiar protons and neutrons, but now have additional energy. The experimental determination of their masses and properties is an important effort at Jefferson Lab. Richards and his colleagues use supercomputers at Oak Ridge National Lab, and the high-performance GPU-enabled (graphics processing unit) clusters at Jefferson Lab, to compute the masses and properties of these excited states from first principles, using lattice QCD. Comparing these calculations with experimental data provides crucial insights into the nature of matter and how the masses of so-called hadronic matter, such as protons and neutrons, arise from QCD. A particularly exciting recent calculation is that of the masses of so-called "exotic mesons," mesons that cannot be constructed from straightforward excitations of a quark and an antiquark, the fundamental building blocks of QCD. The search for such mesons is the aim of the GlueX experiment with CEBAF at 12 GeV. Richards and his colleagues predict that there will be exotic mesons at a mass that will be accessible to GlueX, underpinning the scientific imperative for the experiment. Throughout his career, Richards has received numerous awards, including scholarships at Cambridge and an advanced Fellowship at Edinburgh. He serves on committees such as the Lattice QCD Executive Committee and was the co-organizer of Lattice 2008, the 26th International Symposium on Lattice Field Theory held in Williamsburg, and a panel convener for Forefront Questions in Nuclear Science and the Role of High Performance Computing, held in 2009 in Washington, D.C. |
</li> | </li> | ||
| Line 121: | Line 154: | ||
</li> | </li> | ||
| − | = | + | === Gagik Gavalian === |
| − | + | <li style="display: inline-block;"> | |
| − | + | [[File: gagik_gavalian.jpg|thumb|left|250px| '''Gagik Gavalian: Staff Scientist at Jefferson Lab and Assistant Professor at Old Dominion University.''']] | |
| − | + | '''Dr. Gagik Gavalian''' attended Yerevan State University and graduated in 1996 with a | |
| − | + | major in Physics. He obtained his Ph.D. in Nuclear Physics from the University of | |
| − | + | New Hampshire in May 2004. Gagik then served as a Post Doctoral Research | |
| − | + | Associate at Old Dominion University until 2008. He then assumed the role of | |
| − | + | Assistant Professor at Old Dominion until 2014, where he taught introductory | |
| − | + | physics and conducted research at Jefferson Lab. Gagik played an instrumental | |
| − | + | role in the Hall B data mining efforts leading to multiple publications on studies of | |
| − | + | nuclear effects in electron-nucleus scattering. Gagik joined Jefferson Lab as a staff | |
| − | + | scientist in 2014 and has been working on preparing the CLAS12 data analysis | |
| − | + | packages towards expedient analysis. He also mentors Doctoral candidates and | |
| − | + | college students. For past four years Gagik worked on implementing CLAS12 | |
| − | + | detector reconstruction packages in cloud distributed CLARA framework. CLAS12 | |
| − | + | detector was successfully commissioned in February 2017 with reconstruction | |
| − | + | software successfully tested for full data production. For the past (2017-2018) year | |
| − | + | Gagik was leading effort in development of physics analysis software for CLAS12 | |
| − | + | experimental data. | |
| − | + | </li> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | = Data created with by combining PODM with PATRONS = | ||
<gallery heights=300px widths=300px mode="slideshow"> | <gallery heights=300px widths=300px mode="slideshow"> | ||
File:White_nt_0_1000000.png | File:White_nt_0_1000000.png | ||
| Line 165: | Line 185: | ||
File:White_nt_delta_10_1591_493.png | File:White_nt_delta_10_1591_493.png | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
Latest revision as of 15:57, 12 November 2020
Contents
Overview
Next generation Imaging for Nuclear Femtography is in its very early stages. The complexity from seven-dimensional data and many scales and levels of interactions between the colliding particles and what is observed create many challenges. To address these challenges the “Next-generation imaging filters and mesh-based data representation for phase-space calculations in nuclear femtography (CNF19-04)” project proposed to put together an interdisciplinary team to:
- leverage advanced computational methods and software design approaches from medical imaging for processing phase space distributions in nuclear femtography experiments, with the long term aim to enable next-generation process simulations, data analyses, and physics model comparisons and
- organize a mini-workshop that will: (a) bring world-wide leaders from Imaging in Health Care and (b) nation- and state-wide experts in transferring technology to industry in VA.
Two of the most advanced groups world-wide in Medical Image Computing and Image Guided Neurosurgery are in Brigham and Women's Hospital (BWH), Harvard Medical School. Namely the Surgical Planning Laboratory (SPL) lead by Dr. Ron Kikinis and Image Guided Neurosurgery (IGNS) Laboratory lead by Dr. Alexandra Golby.
- Monday, August 19, the objective is to focus on Lessons Learned from Medical Image Computing at SPL and IGNS at Harvard; together both labs have more than 40 years of experience and many 100’s of person-years of accumulated research.
Job creation and transfer of technology to industry is as crucial for sustaining basic research as the quality of research in general and specifically in Nuclear Femtography. However, nurturing and flexibility as it is demonstrated at the Institute for Advanced Study at Princeton and argued in the “Usefulness of Useless Knowledge”[1] is required.
- Tuesday, August 20 the objective is to focus on Lessons Learned form empirical evidence, address economic strengths and weaknesses of various approaches to generating inventions and innovations, in part address intellectual property and protection --a key part of the process for job creation-- and focus on VA ecosystem, state-based incentives and programs for innovation. Tuesday’s discussion will be lead by Prof. James Koch. Mr. James Cheng and Dr. Steve Pieper.
The CNF Imaging Workshop expected to be highly interactive as participants will transfer know-how from the medical imaging community to basic physics in this case nuclear femtography.
The tessellation data in figure (above) depict a spatial distribution of up quarks as a function of proton's momentum fraction carried by those quarks; bX and bY, spatial coordinates (in 1/GeV = 0.197 fm) defined in a plane perpendicular to the nucleon’s motion, x is the fraction of proton’s momentum and color denotes probability density for finding a quark at given (bX, bY, x). These preliminary data are generated by Dr. Sznajder and processed/tessellated with CRTC's CNF_I2M tool. Their visualization is accomplished by Dr. Gavalian using Paraview.
Schedule
August 19: (Discuss lessons learned from the medical imaging discipline)
- Morning Session: 9:30AM to 12:30 (Chair: Christian)
- Welcome and Logistics for the workshop (Nikos): 9:30AM to 9:40AM
- Introduction to Jefferson Lab (Bob): 9:40AM to 9:55AM
- Brief review of CoS at ODU's efforts with CNF Initiative (Gail): 9:55AM to 10:10AM
- Introduction to Center for Nuclear Femtography (David): 10:10AM to 10:25AM
- Imaging subnuclear systems: Concepts, objectives, applications (Christian): 10:25AM to 10:45AM
- External Speakers: Ron Kikinis and Alexandra Golby: 10:45AM to 12:30PM
- Lunch Break: 12:30PM to 1:30PM
- Afternoon Session: 1:30PM to 3:45PM (Chair: Gagik)
- Focus on software design and building an ecosystem and community around 3D Slicer
- External Speakers:Tina Kapur, Steve Pieper, and Andrey Fedorov
- Dinner at 7:30PM -- TBD
August 20: Broader Impact to the Community
- Morning BoF Session : 10:30AM to 12:00PM (Chair: Nikos)
- Focus on Empirical Data and Lessons Learned on Innovation broadly and more specifically in VA.
- External Speakers: James Koch, James Cheng and Steve Pieper
- Lunch Break: 12:00 to 1:30PM
- Afternoon Session: 1:30PM to 3:30PM (Chair: Christos/Nikos)
- Focus on JLAB projects with concrete examples on imaging CNF data
- Experimental Physicist's Point of View on Imaging for Nuclear Femtography (Gagik): 1:30PM to 2:00PM
- Discussion Moving Forward (Open to all attendees and to be moderated by Nikos): 2:00PM to 3:30PM
- Focus on JLAB projects with concrete examples on imaging CNF data
Registration
Register here: https://cnf_imaging_workshop.eventbrite.com
Presenters
External Visitors
Dr. Ron Kikinis
Dr. Ron Kikinis is the Robert Greenes Distinguished Director of Biomedical Informatics in the Department of Radiology at at Harvard Medical School and "Institutsleiter" of Fraunhofer MEVIS and Honorary Professor of Medical Image Computing at the University of Bremen. He is the founding Director of the Surgical Planning Laboratory, Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, and a Professor of Radiology at Harvard Medical School. This laboratory was founded in 1990. Before joining Brigham & Women's Hospital in 1988, he trained as a resident in radiology at the University Hospital in Zurich, and as a researcher in computer vision at the ETH in Zurich, Switzerland. He received his M.D. degree from the University of Zurich, Switzerland, in 1982. In 2004 he was appointed Professor of Radiology at Harvard Medical School. In 2009 he was the inaugural recipient of the MICCAI Society "Enduring Impact Award". On February 24, 2010 he was appointed the Robert Greenes Distinguished Director of Biomedical Informatics in the Department of Radiology at Brigham and Women's Hospital. On January 1, 2014, he was appointed "Institutsleiter" of Fraunhofer MEVIS and Professor of Medical Image Computing at the University of Bremen. Since then he is commuting every two months between Bremen and Boston. During the mid-80's, Dr. Kikinis developed a scientific interest in image processing algorithms and their use for extracting relevant information from medical imaging data. Since then, this topic has matured from a fairly exotic topic to a field of science. This is due to the explosive increase of both the quantity and complexity of imaging data. Dr. Kikinis has led and has participated in research in different areas of science. His activities include technological research (segmentation, registration, visualization, high performance computing), software system development, and biomedical research in a variety of biomedical specialties. The majority of his research is interdisciplinary in nature and is conducted by multidisciplinary teams. The results of his research have been reported in a variety of peer-reviewed journal articles. He is an author and co-author of over 300 peer-reviewed articles. As part of his service to the community, he serves as reviewer for a large number of journals and conferences from the fields of imaging, biomedical engineering and, computer science. He has served and is serving as member of external advisory boards for a variety of centers and research efforts. He is the Principal Investigator of 3D Slicer, a free open source software platform for image analysis and visualization. Over the years Dr. Kikinis has served as the Principal Investigator(PI) and site PI of a number of large and small NIH and NSF funded grants (see here for his NIH funding). This includes the National Alliance for Medical Image Computing (NA-MIC). He is currently serving as the PI of the Neuroimaging Analysis Center (NAC) and the Quantitative Image Informatics for Cancer Research (QIICR). He is also the Director of Collaborations for the National Center for Image Guided Therapy (NCIGT).
Dr. Alexandra Golby
Dr. Alex Golby's is the is Haley Distinguished Chair in the Neurosciences and Professor of Neurosurgery & Radiology at Harvard Medical School. She focuses on the translation of a broad range of neuroimaging techniques to neurosurgical planning and intraoperative guidance. The overarching goal of this work is to help surgeons perform optimal brain surgery by defining and visualizing critical brain structures and pathologic tissue to be removed. Since 1998, she has worked on the development and validation of fMRI for the pre-operative evaluation of patients with lesions in and near critical areas of the brain. This has been a translational research effort which adapted fMRI, initially developed as a neuroscience technique to be applied in groups of subjects to make statistical inferences about populations, to the vastly different scenario of clinical decision-making for individual patients. Since her research program began at BWH in 2003,Dr. Alexandra Golby's group has developed new techniques for the use of fMRI in single subject analyses necessary for surgical planning. In addition, they have developed numerous acquisition strategies geared towards accommodating the limited neurologic functions of some patients as well as analytic approaches to maximize the utility of fMRI for surgical planning. Presurgical fMRI has the potential to bring meaningful pre-operative individualized functional anatomy mapping to neurosurgeons around the world as an alternative to awake mapping, a technique which is demanding and remains limited to very specialized centers. Dr. Alexandra Golby has also worked extensively on the translation of diffusion MRI (dMRI) including tensor imaging (DTI) to map white matter anatomy in neurosurgical patients. Diffusion MRI allows the in vivo depiction of the location, course and integrity of macroscopic white matter tracts in the brain through a process known as tractography. As with fMRI, the translation of this technology to clinical decision-making has required numerous fundamentally novel approaches. Her group has developed segmentation approaches for defining tracts based on high dimensional clustering as well as statistical atlases which allow labeling of individual patient tracts even in the setting of mass effect and peritumoral edema. Her group works collaboratively with MRI physics and MRI analysis groups to continue to be at the forefront of technical innovation. They have released many of our tools to the public via 3D Slicer (slicer.org) and have organized several international challenge workshops to apply diffusion techniques to real world clinical data. With both these methods, translation of the technology required understanding of clinical needs, constraints, and opportunities for improved clinical care. Specific analysis techniques needed to be developed to adopt these techniques so that they were applicable to single subject data, and, in particular, to neurologic patients who have structural lesions and often are limited by their neurological deficits. In these efforts, they work closely and collaboratively with scientists in radiology and computer science to translate emerging technical innovations into the operating room. Another major area of translational investigation is in the development of intraoperative imaging techniques. Dr. Golby was the lead surgeon in developing the AMIGO (Advanced Multi-modality Image-Guided Operating Suite) at BWH and serve as the Co-director of AMIGO. AMIGO is one of the key resources of the National Center for Image Guided Therapy funded by NIH. This suite contains all contemporary imaging methods within an operating room environment and was specifically designed to support translational research. The suite is the site of many of surgical procedures in which we are developing strategies for intraoperative imaging and guidance. Several of her group's important research efforts are built on the AMIGO platform. These include the intra-operative use of high field MRI including development of intra-operative dMRI tractography. They have also leveraged the resources of the AMIGO suite to develop novel strategies to simplify intraoperative imaging using techniques such as ultrasound and stereovision to give surgeons information in near real time to guide surgery. Another area of research leveraging the resources of AMIGO is the development of tissue level molecular imaging. Dr. Alexandra Golby's group has funded collaborative projects using mass spectrometry, Raman spectroscopy, and fluorescence imaging. Their eventual goal is to give surgeons in most settings tools that will help them to perform safer and more effective surgery.
Mr. James Cheng
James Cheng (MBA ’87) is is former Virginia’s secretary of commerce and trade; he managed 13 agencies, including economic development, export, tourism, small business, housing, mining and energy. Currently an angel investor and entrepreneur with more than 20 years of experience in information technology and government contracting. He has adopted another title according to his LinkedIn profile: Chief Balancing Officer. Cheng and his wife, Jeanette Wang, are raising their first child, 3-year-old Joshua. In addition to fatherhood, Cheng juggles five part-time jobs, ranging from advisor to investor to adjunct professor. He is very fortunate to not have just one job. From 2010 until 2014, Cheng worked as Virginia’s secretary of commerce and trade, managing 13 agencies, including economic development, export, tourism, small business, housing, mining and energy. Cheng never imagined he would go into politics as a computer science major from Old Dominion University. He was always a tech guy, and after graduation, he joined his father’s company. In 1994, Mr. Cheng decided to create his own firm. Cheng grew Computer & Hi-tech Management Inc. from five employees to 550 employees, with annual revenues skyrocketing from $300,000 to $90 million by the time he sold it 11 years later. The key to entrepreneurship, as Cheng tells his students, is to solve a pain point. “It might be something they don’t even know exists,” he said. As technology advances, Cheng is confident that it will enable all of us to achieve more balance in our lives. He is always looking for the next greatest thing, and “Balance is it.” Cheng says the skills he gained at the University of Virginia Darden School of Business have been the cornerstone of his success, so it only seemed right to him to create the $1 million James Su-Ting and Jeannette Wang Cheng Scholarship Fund to support students from East Asia. A Principal Donor who served on the Darden School Foundation Board of Trustees from 2008 until the summer of 2015, Cheng lives in Charlottesville with his wife and son. He is currently working on a new angel fund for University of Virginia alumni called CAV Angels.
Dr. Tina Kapur
Dr. Tina Kapur is Executive Director of Image Guided Therapy in the Department of Radiology at Brigham and Women's Hospital and Harvard Medical School. Her research interests and accomplishments are in the area of medical image computing and computer aided interventions in image-guided neurosurgery, surgical navigation, and MR-guided pelvic brachytherapy. She has numerous publications in medical image segmentation, and is the holder of several issued US and international patents in the field of surgical navigation. She leads national and international outreach and collaboration efforts for image-guided therapy and is particularly interested in fostering collaborations between efforts in open science to accelerate important discoveries that improve health and save lives. She is the founding director for the first open-science hackathons for medical image computing, "NA-MIC Project Week", which began in 2005, and has been running continuously (twice a year) since then. She received her Ph.D. in Electrical Engineering and Computer Science from the Massachusetts Institute of Technology in 1999. She was the Chief Scientist at a Boston area surgical navigation company, Visualization Technology Inc., and upon its acquisition by GE Healthcare, the Chief Scientist at the GE Navigation.
Dr. Steve Pieper
Dr. Steve Pieper Founder and CEO of Isomers,Inc. He is a Computer Scientist with a long-time interest in applying Computer Graphics to problems in Medical Imaging, Surgical Planning, and Biomechanics Simulation expressed through entrepreneurship and academic research. He received his Computer Science degree from UC Berkeley and M.S. and Ph.D. from the MIT Media Lab. At the SPL, Steve is the Engineering Core PI on the Neuroimage Analysis Center. He is also responsible for application engineering and dissemination activities in the National Alliance for Medical Image Computing. He has also taken on responsibility for the ongoing architectural development and maintenance of the 3D Slicer, the SPL's flagship software system. Currently, Steve is actively working on Version 4 of 3D Slicer (Slicer4). Steve is active in a variety of research collaborations related to software development, surgical applications, high performance computing, facial animation, haptics, registration and segmentation. His thesis topic is Computer-Aided Plastic Surgery. He is a Faculty in Engineering at Dartmouth College and an adjunct teacher of Computer Science and Computer Graphics at Berkeley and MIT. Dr. Pieper is the founder, former President, CTO and Board Member at Medical Media Systems. He is the current founder and CEO of Isomics,Inc. He has Eight+ issued US Patents and has managed medical software from development through FDA clearance to reimbursement and clinical acceptance.
Dr. Andrey Fedorov
Dr. Andrey Fedorov is an Assistant Professor in Radiology at the Surgical Planning Laboratory (SPL), Department of Radiology, Brigham and Women's Hospital and Harvard Medical School. Andrey is former member of CRTC group and he moved SPL in 2009 after obtaining his PhD in Computer Science from The College of William and Mary in Virginia. His research is in translation and validation of medical image computing technology in clinical research applications, with the focus on quantitative imaging, imaging informatics and image-guided interventional procedures. Andrey is committed to advancing the role of reproducible science, data sharing and open source software in academic research. He has contributed to a number of open source projects, most notably 3D Slicer (http://slicer.org). Together with Ron Kikinis, he is a co-PI of the Quantitative Image Informatics for Cancer Research (QIICR) project (http://qiicr.org) focused on developing open source informatics technology in support of quantitative imaging biomarker development, and interoperable sharing of the imaging biomarker data using the Digital Imaging and Communications in Medicine.
ODU/JLAB
James Koch
James V. Koch is Board of Visitors Professor of Economics Emeritus and President Emeritus at Old Dominion University. He has held research and teaching positions at Illinois State University, California State University at Los Angeles, the University of Grenoble (France), Brown University, Rhode Island College, Ball State University, the University of Hawaii, the Royal Melbourne Institute of Technology in Australia and the University of Montana. He served as President of the University of Montana (1986-1990) and Old Dominion University (1990-2001) and was named one of the 100 most effective college presidents in the United States. Dr. Koch’s research on the risk-taking behavior of corporate CEOs was funded by the Kauffman Foundation and was published as Born, Not Made (Praeger, 2008, co-author James L. Fisher). Another book---America For Sale, a study of the purchase of U.S. assets by foreigners (Praeger, Fall 2009, co-author, Craig T. Bouchard)---earned wide attention, especially in the steel industry, its primary focus. Dr. Koch served as a member of the board of the Wheeling-Pittsburgh Steel Company and its successor, Esmark. Dr. Koch has published twelve books and 110 articles in refereed journals. His research has focused primarily upon applied microeconomics topics and his work on the economics of intercollegiate athletics, the economics of discrimination and affirmative action, TQM, and the economics of education has been reprinted and cited frequently. He has done extensive work in the economics of e-commerce and has served as a consultant/expert witness for more than 100 legal firms, corporations, and universities. Beginning in 2015, Dr. Koch began to produce a State of the Commonwealth Report that focuses on Virginia economic issues. For eighteen years, he has produced the widely cited State of the Region Report for the 1.7 million population Hampton Roads, Virginia, metropolitan area. James Koch has led or been a part of teams commissioned by boards of trustees of more than 50 universities to evaluate the strategic positions of these institutions. He has evaluated the presidents of several institutions including the University of Hawaii, Muhlenberg College, Talladega College, Louisiana Tech, the University of Central Florida, Auburn University, and the University of Alaska. His 1996 book, Presidential Leadership (co-authored with James L. Fisher) is used as a reference and textbook in many universities and leadership institutes.
Gail Dodge
Dr. Gail Elizabeth Dodge is Dean of the College of Sciences and Professor of Physics at Old Dominion University. She has been a member of the faculty since 1995. She received her B.A. from Princeton University and her M.S. and Ph.D. from Stanford University. She was chair of the ODU Department of Physics for six years. In 2015, she was selected by the State Council of Higher Education for Virginia for an Outstanding Faculty Award. Dr. Dodge is an experimental nuclear physicist, studying the protons and neutrons which make up the atomic nucleus. She uses the high energy electron beam at the Thomas Jefferson National Accelerator Facility in Newport News, Virginia, which functions like an enormous electron microscope. Her research centers on the study of the structure of the nucleon, and in particular its intrinsic angular momentum, known as spin. The contribution of the quarks to the nucleon spin has been found to be surprisingly low, around 25%, in high energy lepton scattering experiments. At Jefferson Lab the energy of the electron beam is ideal for investigating the excited states of the nucleon, or resonances. Very little data exist on the spin structure of the nucleon in this energy regime. Gail Dodge she carries out these experiments to probe the underlying quarks and gluons that make up the protons and neutrons. Dr. Dodge served a two-year term as a program manager at the National Science Foundation (NSF) in Arlington, Virginia. There she was responsible for the $17 million experimental nuclear physics program and for coordination with the Office of Nuclear Physics at the Department of Energy (DOE). Prior to working at NSF, she served as a member of the Nuclear Science Advisory Committee, which advises DOE and NSF on priorities for nuclear science funding in the United States.
Robert McKeown
Dr. Robert D. McKeown has been the deputy director for science at Jefferson Lab since May 2010. He also serves as a Governor's Distinguished Continuous Electron Beam Accelerator Facility (CEBAF) Professor at The College of William and Mary. He first became interested in experimental nuclear physics while an undergraduate student at Stony Brook University in Stony Brook, NY, where he received a B.S. in physics in 1974. He then continued his studies at Princeton University, where he received a Ph.D. in 1979. Robert McKeown's research interests include studies of weak interactions in nuclei, neutrino oscillations, parity-violating electron scattering, and the electromagnetic structure of nuclei and nucleons. After one year as a scientist at Argonne National Laboratory, McKeown took a position as assistant professor of physics at the California Institute of Technology. He became an associate professor in 1986 and a professor in 1992. He remained at the California Institute of Technology until joining Jefferson Lab. Over his career, McKeown's exceptional work has been recognized with many awards. He received a National Science Foundation Presidential Young Investigator award in 1984, was the Alexander M. Cruickshank Lecturer at the 1999 Gordon Conference on Nuclear Physics, and is a Fellow of the American Physical Society. He has served on the Nuclear Science Advisory Committee, the Physical Review C editorial board, and on advisory committees for Brookhaven National Laboratory, Fermilab and Jefferson Lab, where he served as chair of the JLab Users Group Board of Directors in 1990-91. In 2009, he received the Tom W. Bonner Prize in Nuclear Physics from the American Physical Society for "his pioneering work on studying nucleon structure using parity-violating electron scattering, in particular for the first measurement of the strange quark contribution to the electromagnetic structure of the proton."
David Richards
Dr. David Richards is Theoretical and Computational Physics at DOE's Jefferson Lab. Richards came to Jefferson Lab as a staff scientist and joint faculty member at Old Dominion University in 1999. He became a full-time staff scientist in 2002 and served as acting Theory Center leader from September 2009 through October 2010. He was appointed deputy director of the Theory Center in mid-October 2010. Richards' current research focus is aimed at garnering a better understanding of so-called "excited states." These are subatomic particles that were once the familiar protons and neutrons, but now have additional energy. The experimental determination of their masses and properties is an important effort at Jefferson Lab. Richards and his colleagues use supercomputers at Oak Ridge National Lab, and the high-performance GPU-enabled (graphics processing unit) clusters at Jefferson Lab, to compute the masses and properties of these excited states from first principles, using lattice QCD. Comparing these calculations with experimental data provides crucial insights into the nature of matter and how the masses of so-called hadronic matter, such as protons and neutrons, arise from QCD. A particularly exciting recent calculation is that of the masses of so-called "exotic mesons," mesons that cannot be constructed from straightforward excitations of a quark and an antiquark, the fundamental building blocks of QCD. The search for such mesons is the aim of the GlueX experiment with CEBAF at 12 GeV. Richards and his colleagues predict that there will be exotic mesons at a mass that will be accessible to GlueX, underpinning the scientific imperative for the experiment. Throughout his career, Richards has received numerous awards, including scholarships at Cambridge and an advanced Fellowship at Edinburgh. He serves on committees such as the Lattice QCD Executive Committee and was the co-organizer of Lattice 2008, the 26th International Symposium on Lattice Field Theory held in Williamsburg, and a panel convener for Forefront Questions in Nuclear Science and the Role of High Performance Computing, held in 2009 in Washington, D.C.
Latifa Elouadrhiri
Dr. Latifa Elouadrhiri is a Senior Staff Scientist in Thomas Jefferson National Accelerator Facility' s Hall B. She received her Ph.D. from the University of Clermont- Ferrand (France) in 1992 for research on the nucleon axial form factor. In 1994 Elouadrhiri came to Jefferson Laboratory for a joint appointment with CNU, joining the lab as a staff scientist in 2001. She was one of the first female staff scientists in the physics division, where she led the measurement of the first observation of electron beam asymmetries in polarized, exclusive Deeply Virtual Compton Scattering. From 2006 to 2015, Elouadrhiri was the Project Manager for the Hall B 12 GeV Upgrade, including all detectors, magnets and infrastructure for the CLAS12 system. Latifa was part of a small group that led the pioneering CLAS paper on the first observation of electron beam asymmetries in polarized exclusive deeply virtual Compton scattering (DVCS). Additionally, she is the spokesperson of the Deeply Virtual Compton Scattering (DVCS) experiment, studying Generalized Parton Distributions. Prior to coming to JLab Latifa performed experiments at Saclay (France) and PSI (Switzerland). Since 2005 Latifa has been the Control Account Manager for the Hall B 12 GeV Upgrade that includes all detectors and magnets for the CLAS12 system. She is a member of the APS Division of Nuclear Physics and was elected APS Fellow in 2011. She has served as member of Nuclear Science Advisory Committee (NSAC), Committee of Visitors (COV). She also served on review panels for funding proposals submitted to the German Research Association (DFG). Latifa Elouadrhiri served as member on several Ph.D. thesis committees, and is the contact person and spokesperson of DVCS experiments to study the Generalized Parton Distributions (GPDs) program with the CLAS12 detector.
Gagik Gavalian
Dr. Gagik Gavalian attended Yerevan State University and graduated in 1996 with a major in Physics. He obtained his Ph.D. in Nuclear Physics from the University of New Hampshire in May 2004. Gagik then served as a Post Doctoral Research Associate at Old Dominion University until 2008. He then assumed the role of Assistant Professor at Old Dominion until 2014, where he taught introductory physics and conducted research at Jefferson Lab. Gagik played an instrumental role in the Hall B data mining efforts leading to multiple publications on studies of nuclear effects in electron-nucleus scattering. Gagik joined Jefferson Lab as a staff scientist in 2014 and has been working on preparing the CLAS12 data analysis packages towards expedient analysis. He also mentors Doctoral candidates and college students. For past four years Gagik worked on implementing CLAS12 detector reconstruction packages in cloud distributed CLARA framework. CLAS12 detector was successfully commissioned in February 2017 with reconstruction software successfully tested for full data production. For the past (2017-2018) year Gagik was leading effort in development of physics analysis software for CLAS12 experimental data.