Difference between revisions of "Medical Imaging Example Meshes"
Spyridon97 (talk | contribs) (→COVID-19-Main-Protease-6y2e) |
Spyridon97 (talk | contribs) (→COVID-19-Main-Protease-6y2e) |
||
| Line 46: | Line 46: | ||
==COVID-19-Main-Protease-6y2e== | ==COVID-19-Main-Protease-6y2e== | ||
* Input image : Dimensions () with spacing () | * Input image : Dimensions () with spacing () | ||
| − | * Uniform with Delta = 0.5: | + | * Uniform with Delta = 0.5: 2,509,202 tetrahedra |
<gallery mode="packed" heights=250px> | <gallery mode="packed" heights=250px> | ||
Revision as of 05:56, 19 March 2020
Contents
3D Example Meshes
The directory containing the 3D input data is located in the 3D folder of Medical_Imaging_Data.
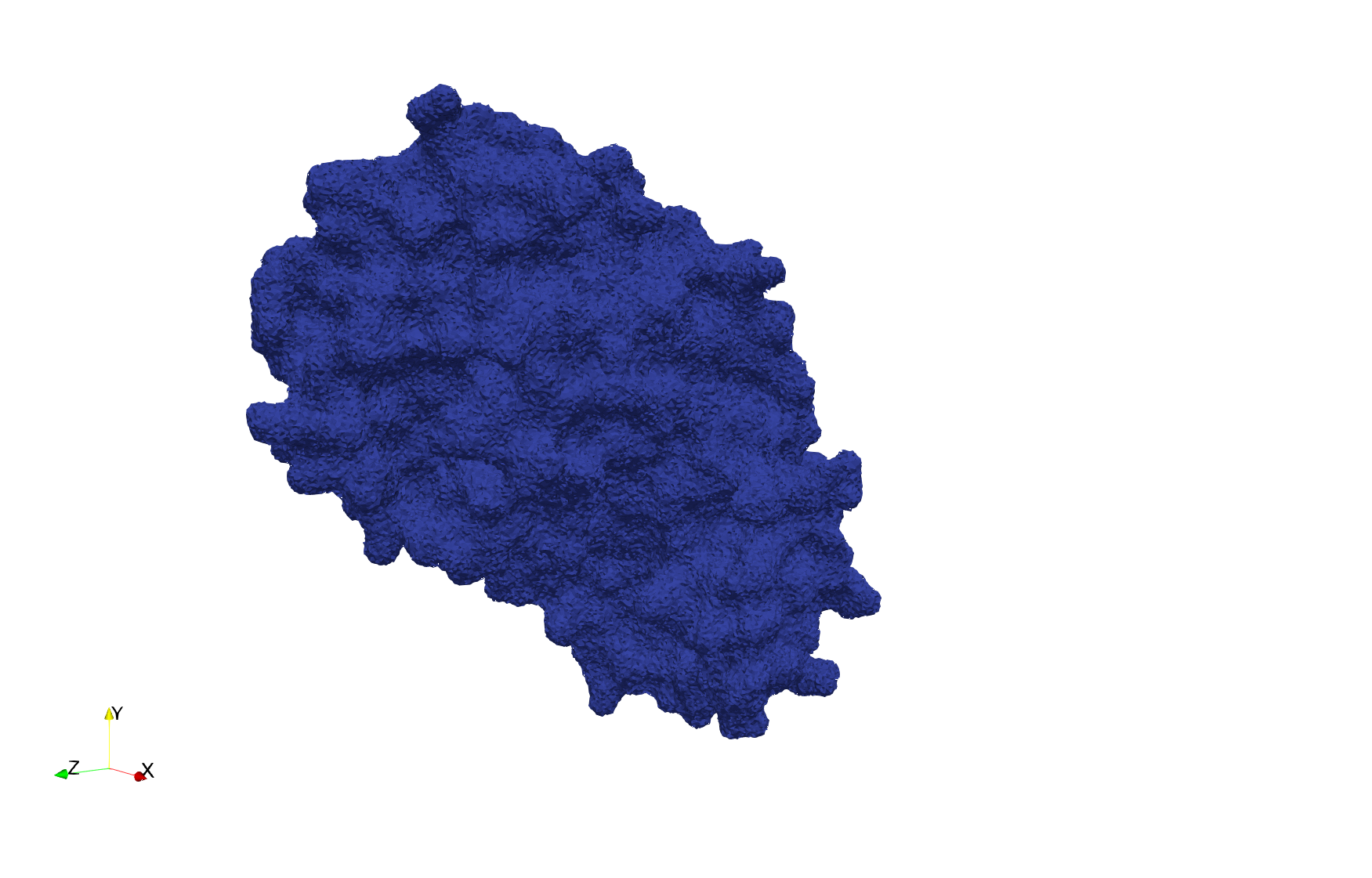
COVID-19-Spike-Glycoprotein-6vxx
- Input image : Dimensions () with spacing ()
- Uniform with Delta = 0.5: tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vxx.nrrd --delta 0.5 --output ./COVID-19-Spike-Glycoprotein-6vxx,d=5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 spike glycoprotein. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
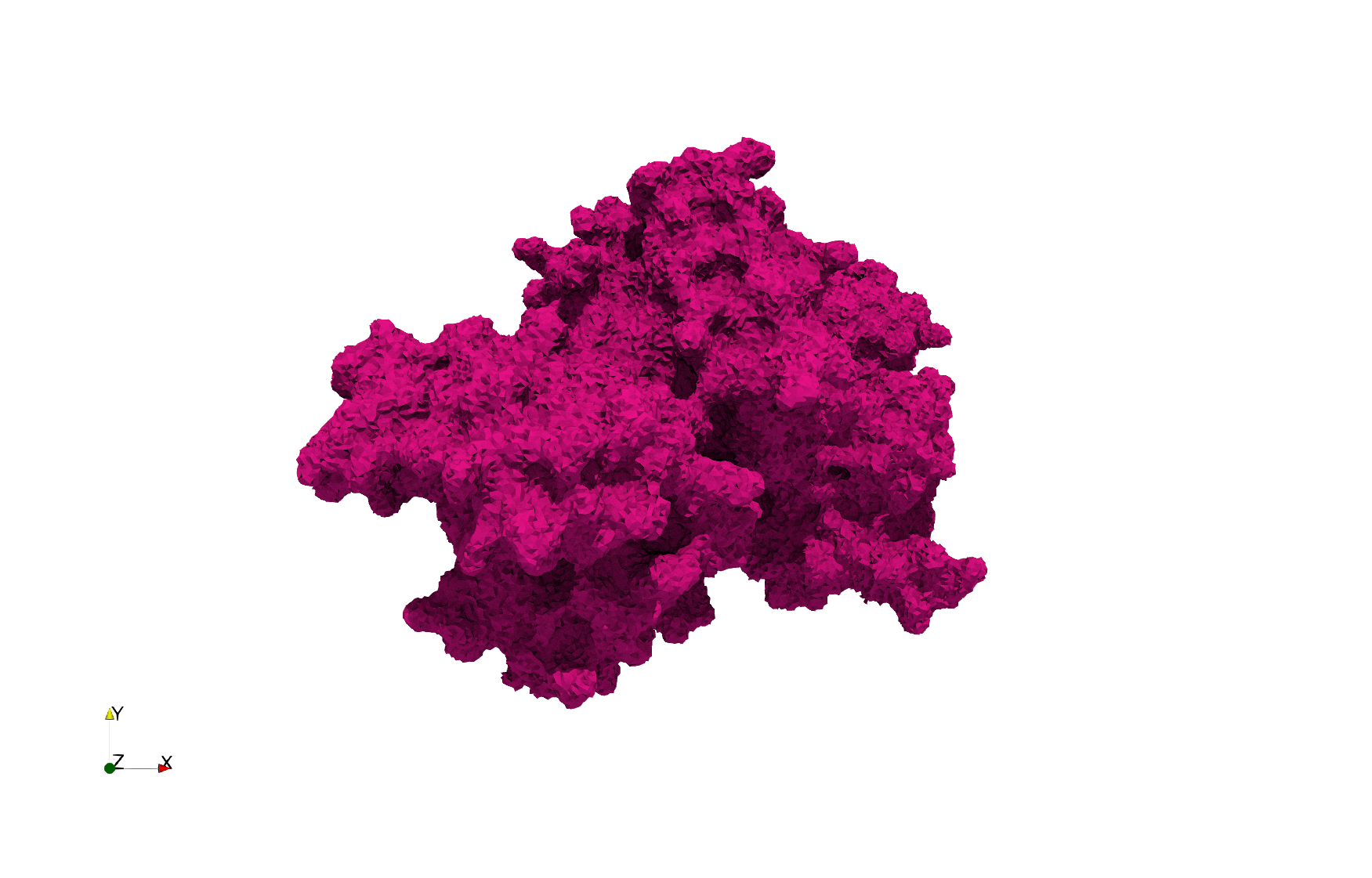
COVID-19-NSP-15-Endoribonuclease-6vww
- Input image : Dimensions () with spacing ()
- Uniform with Delta = 0.3: tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.3: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-NSP-15-Endoribonuclease-6vww.nrrd --delta 0.3 --output ./COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 NSP15 Endoribonuclease. Biological assembly data was retrieved from the molecule 6vww, Protein Data Bank.
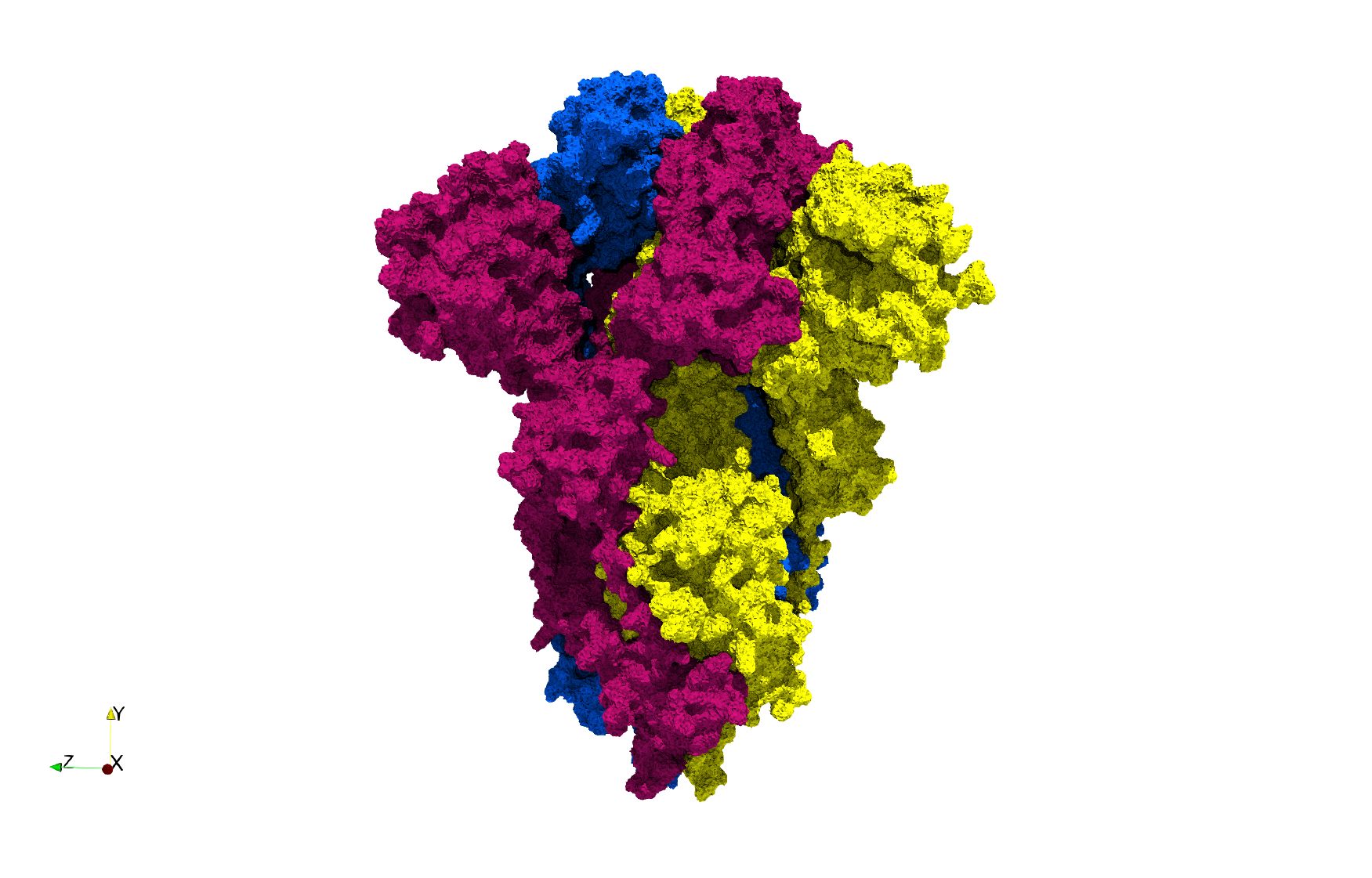
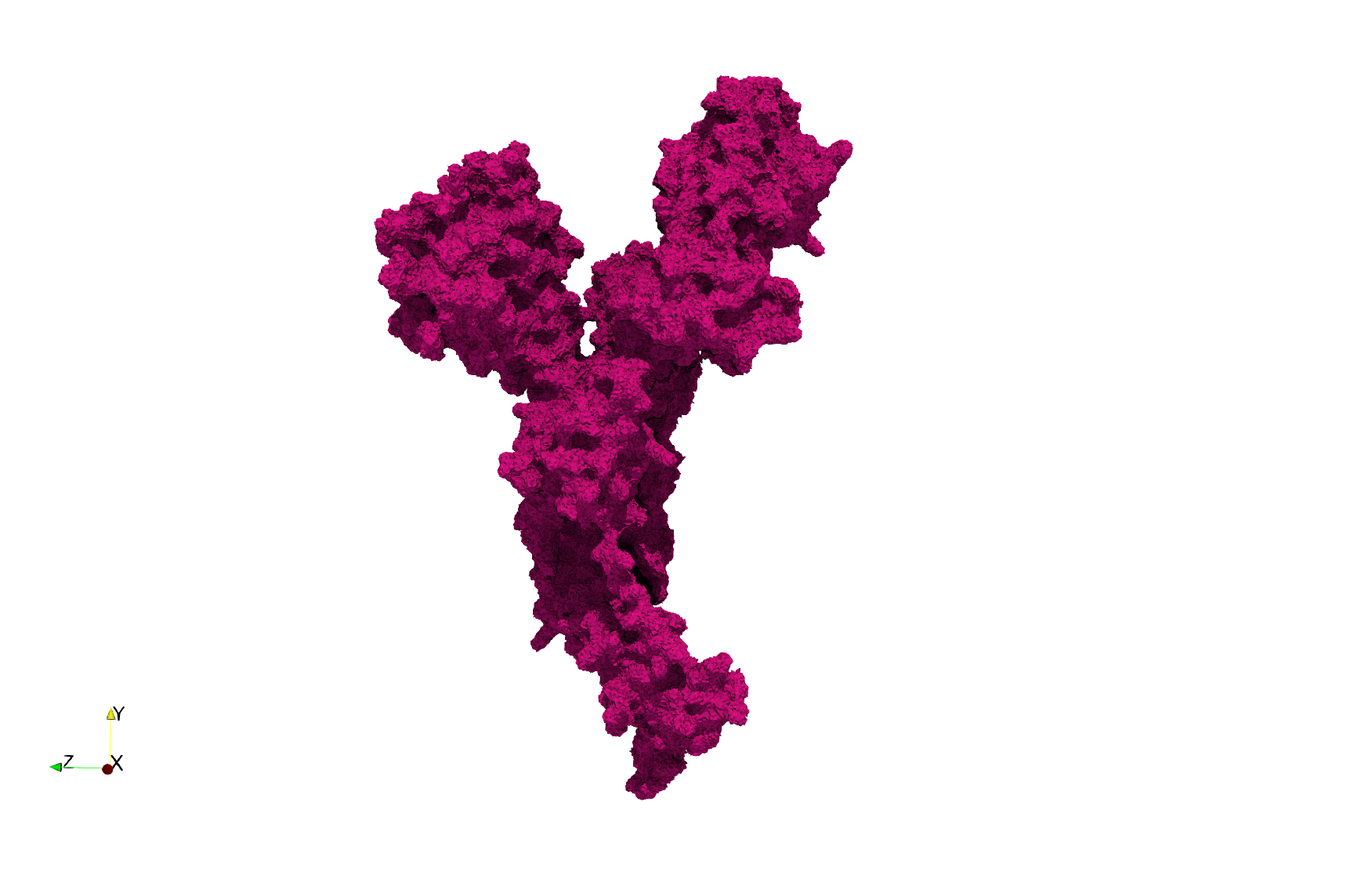
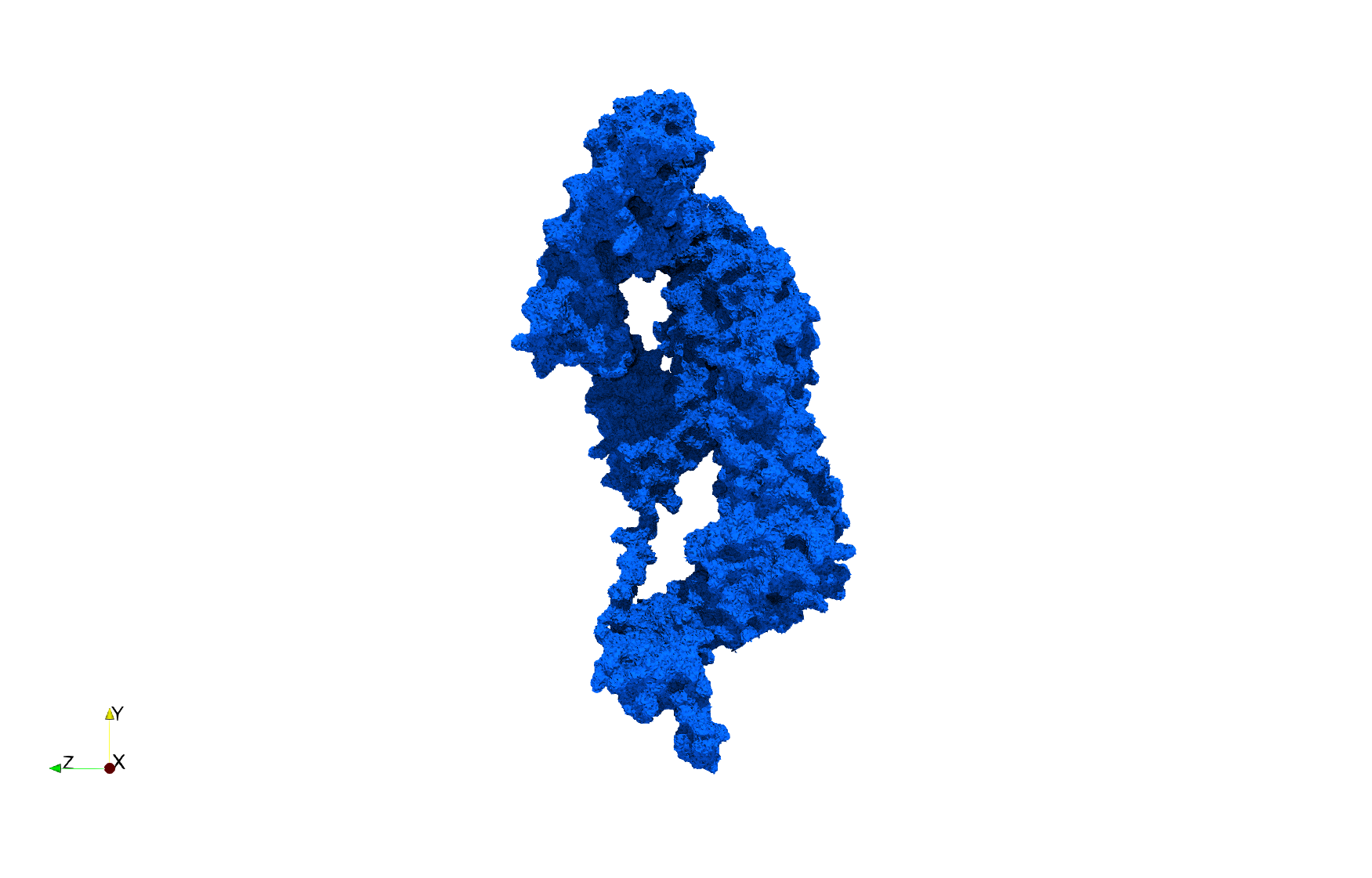
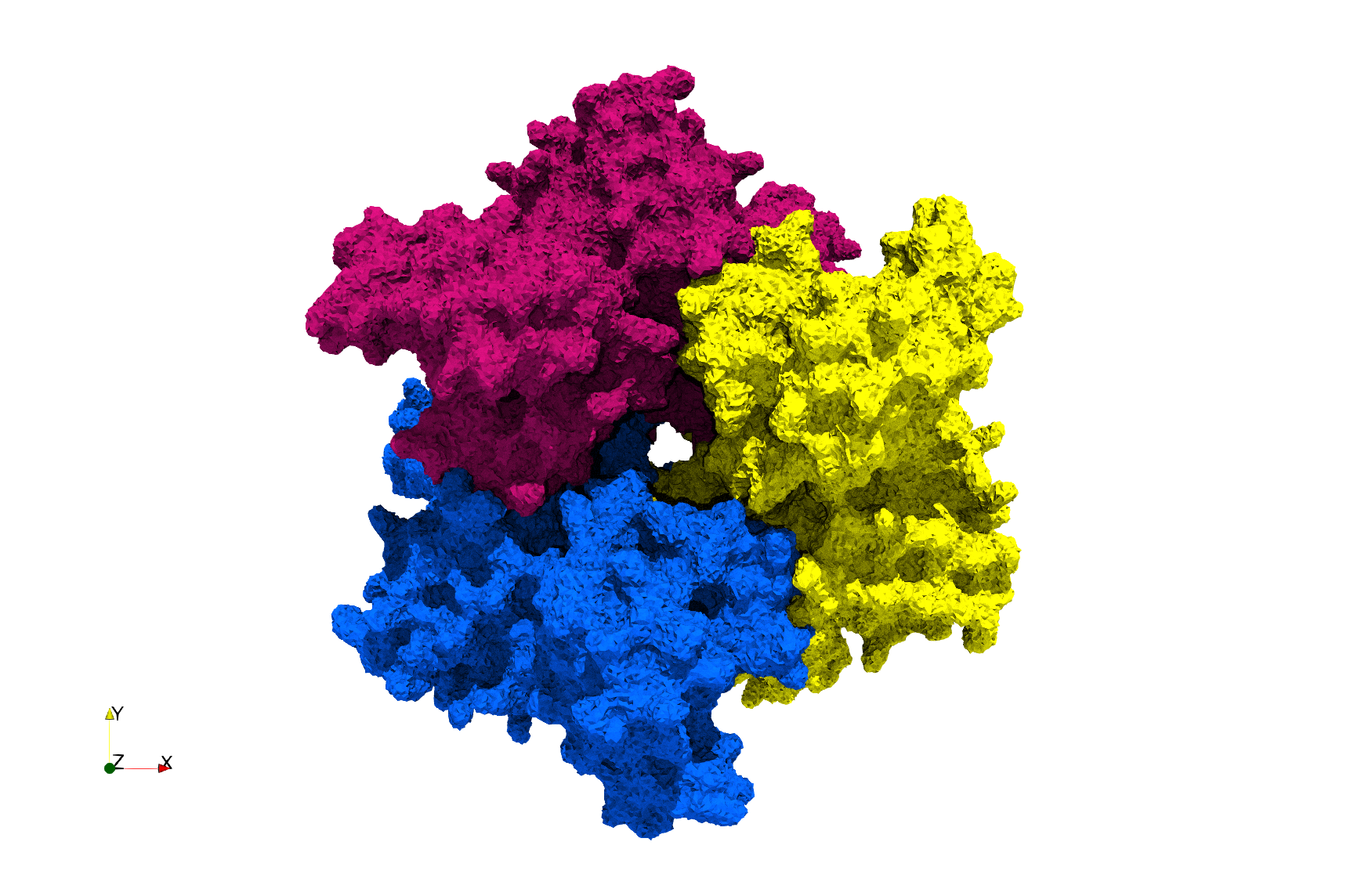
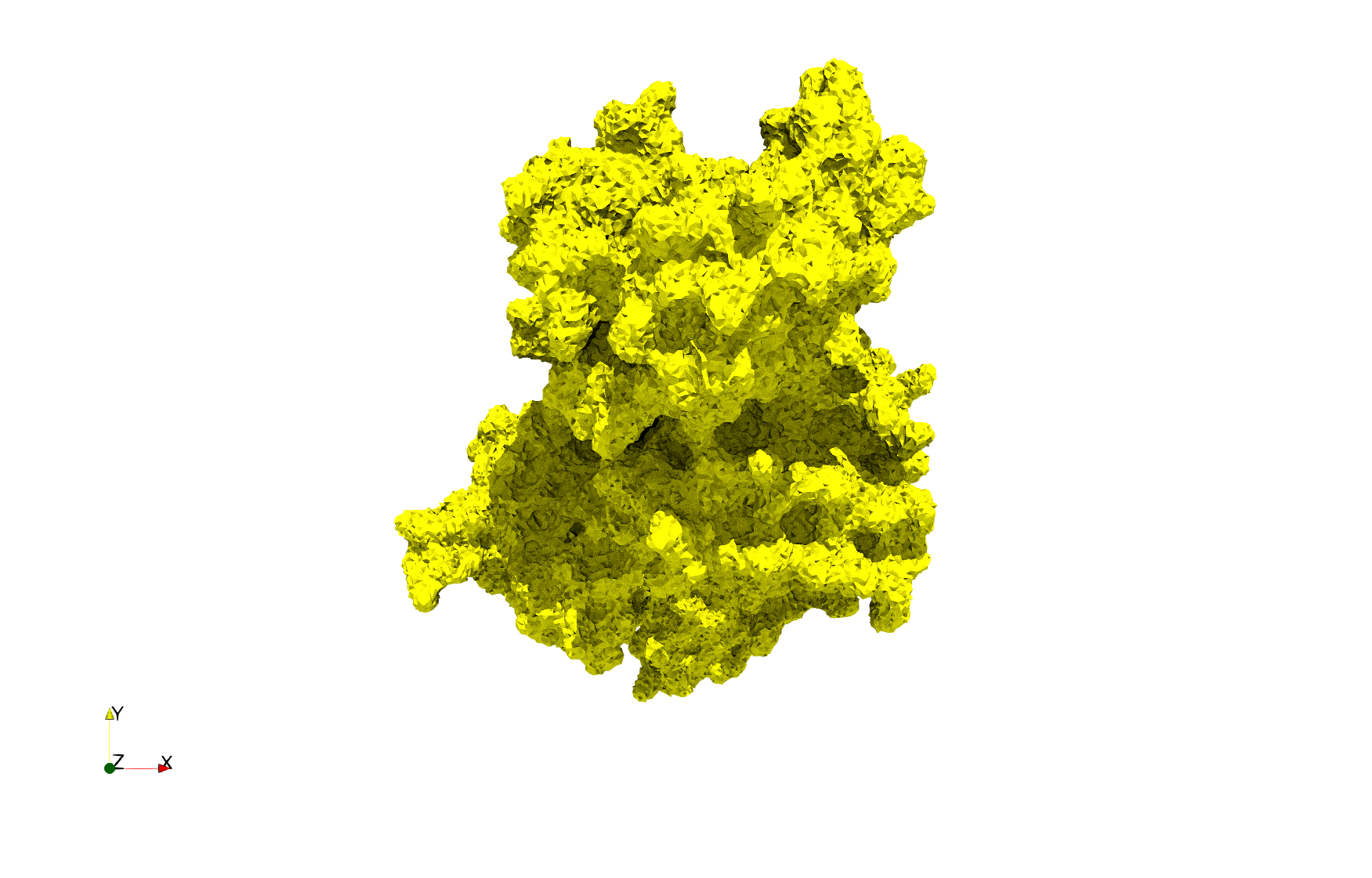
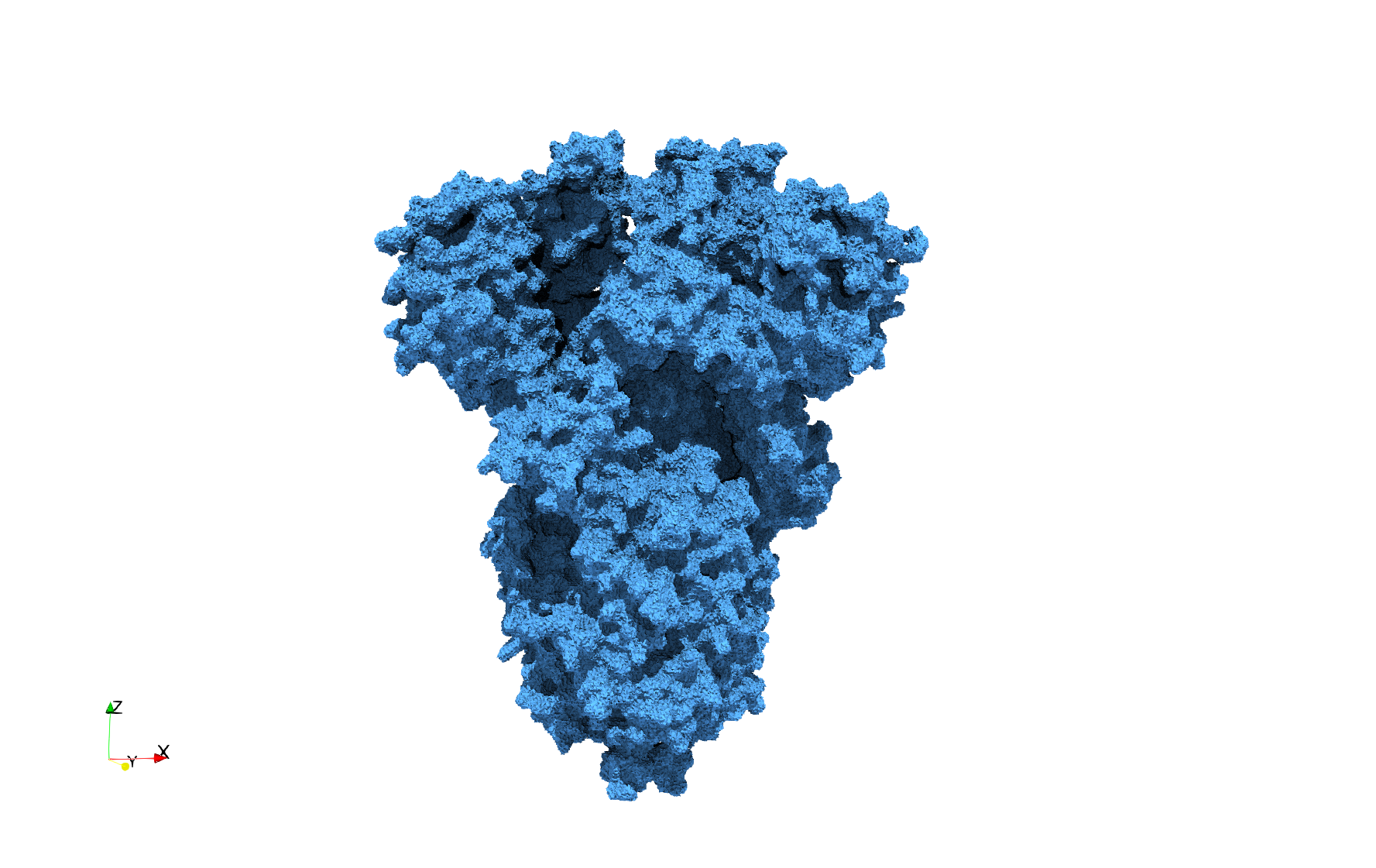
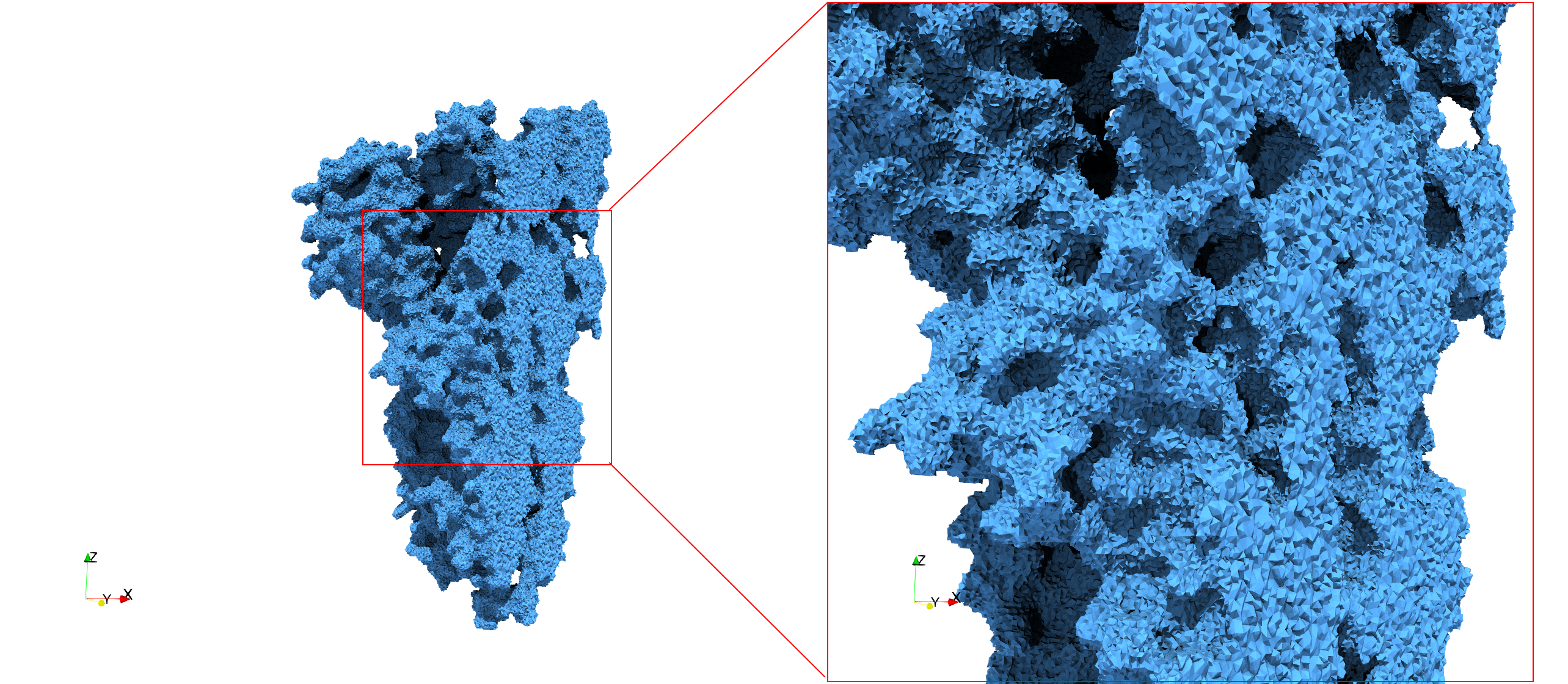
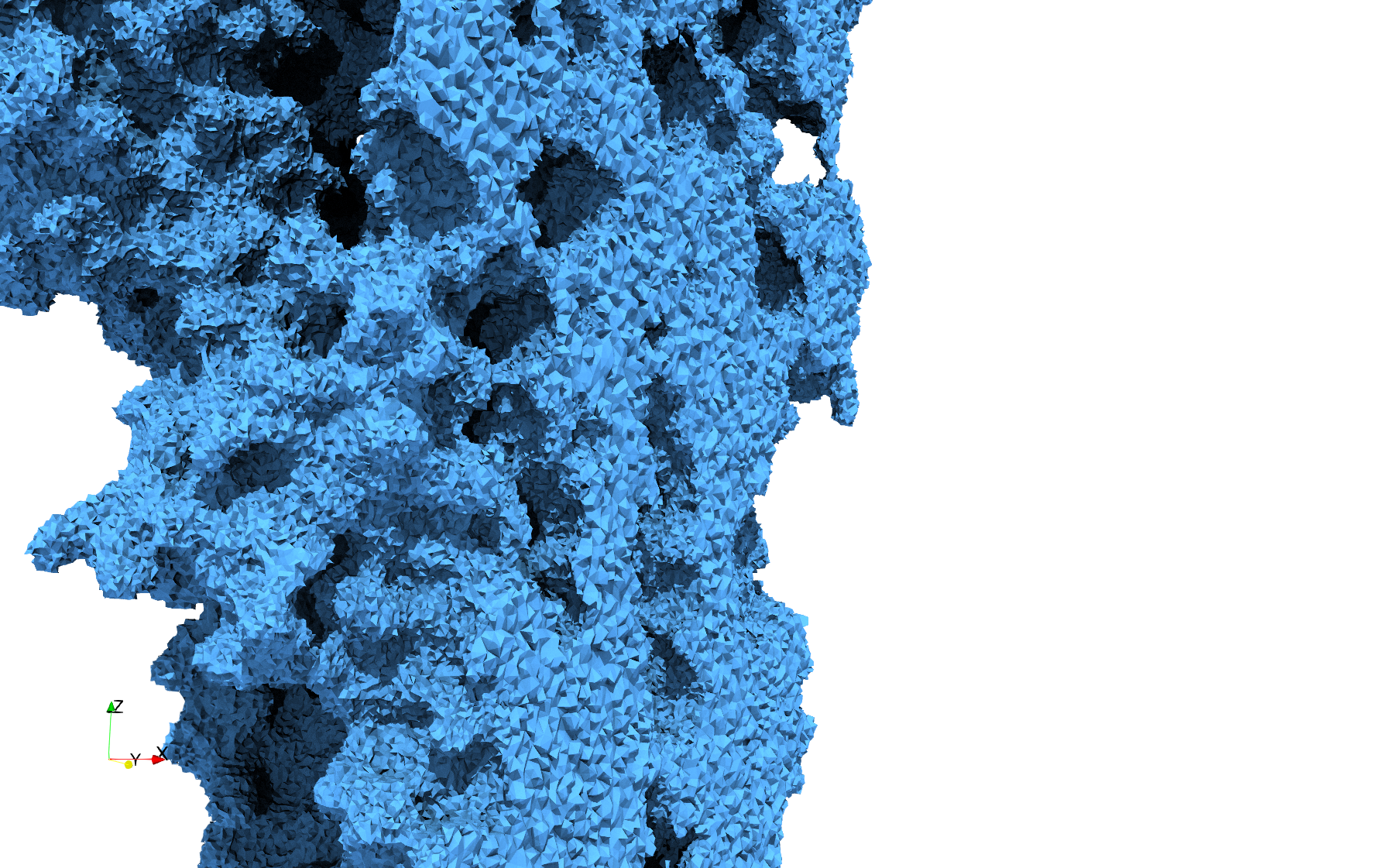
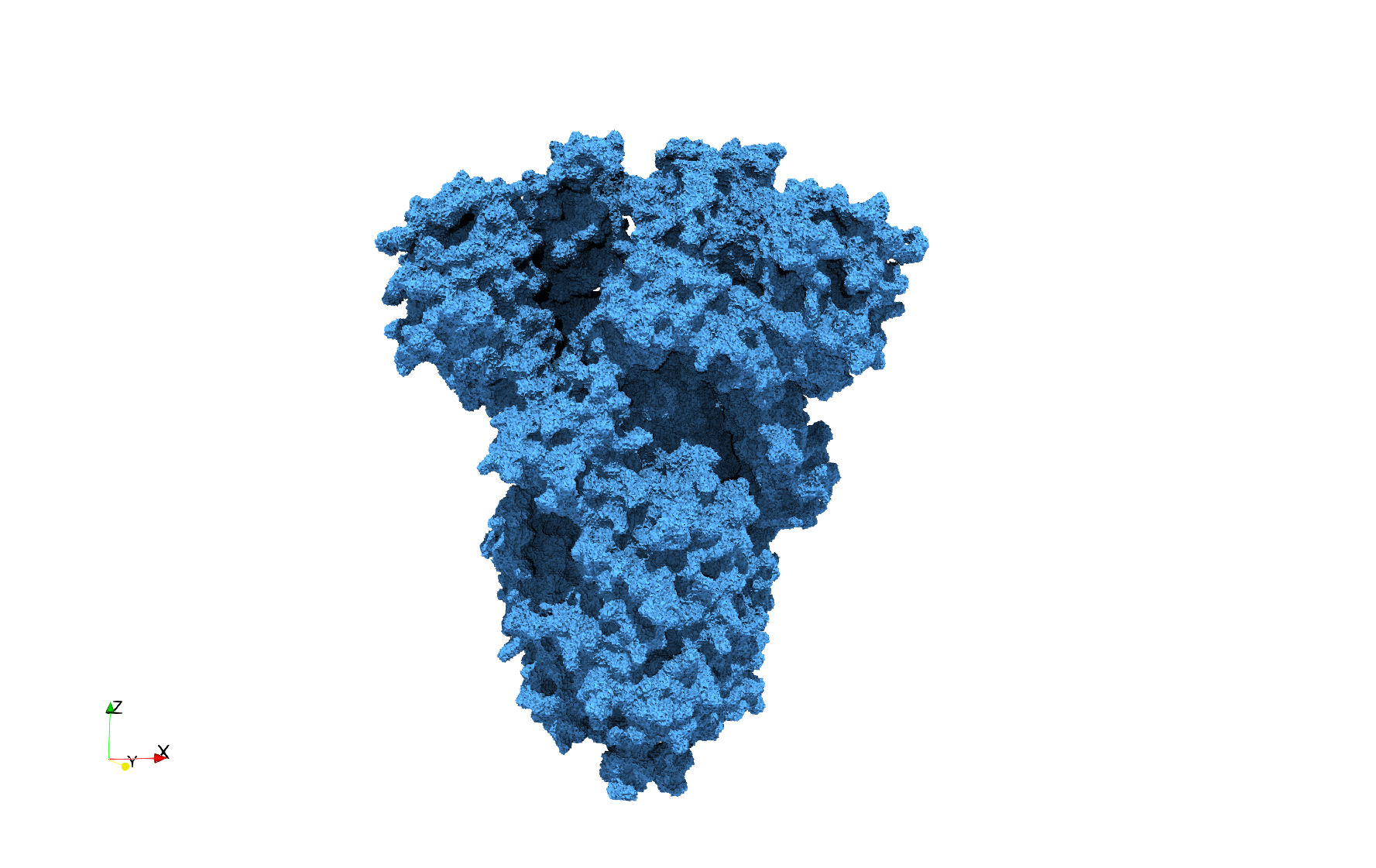
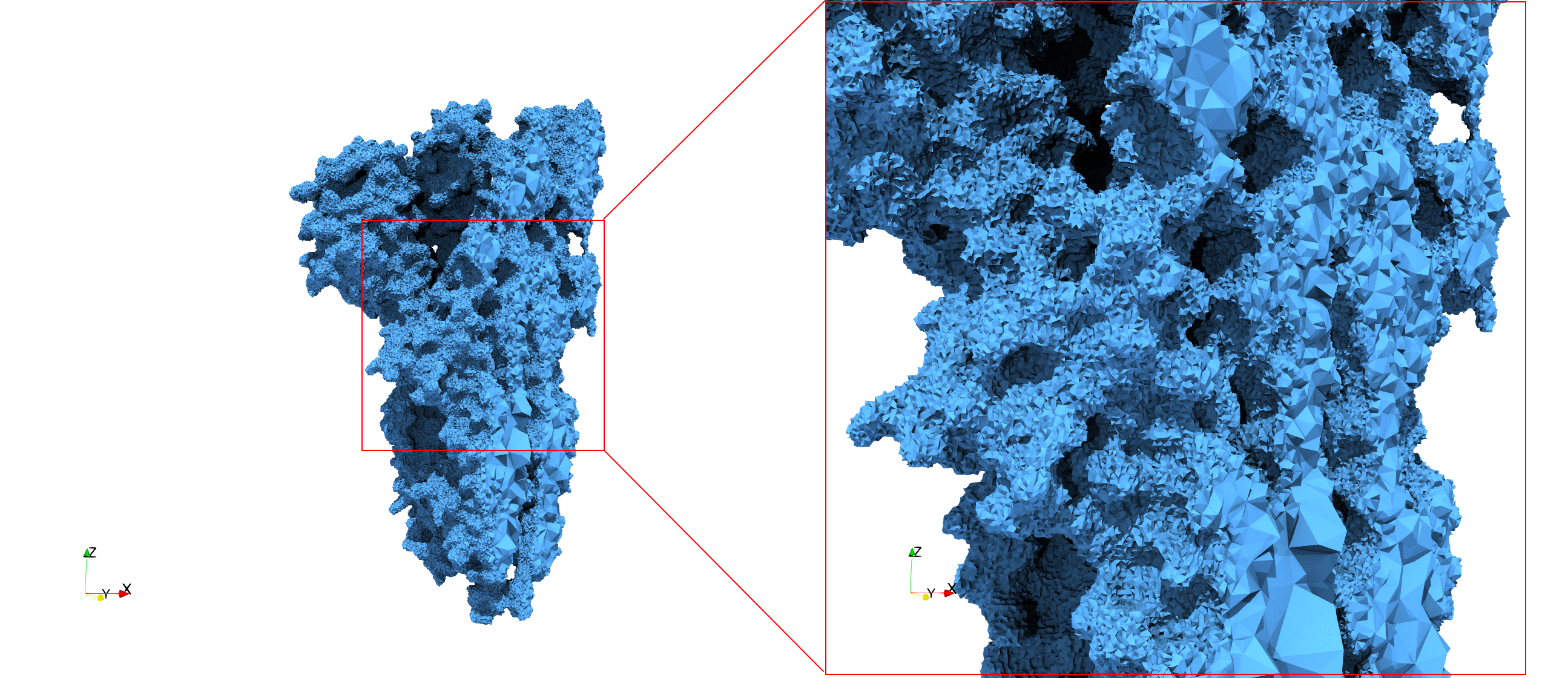
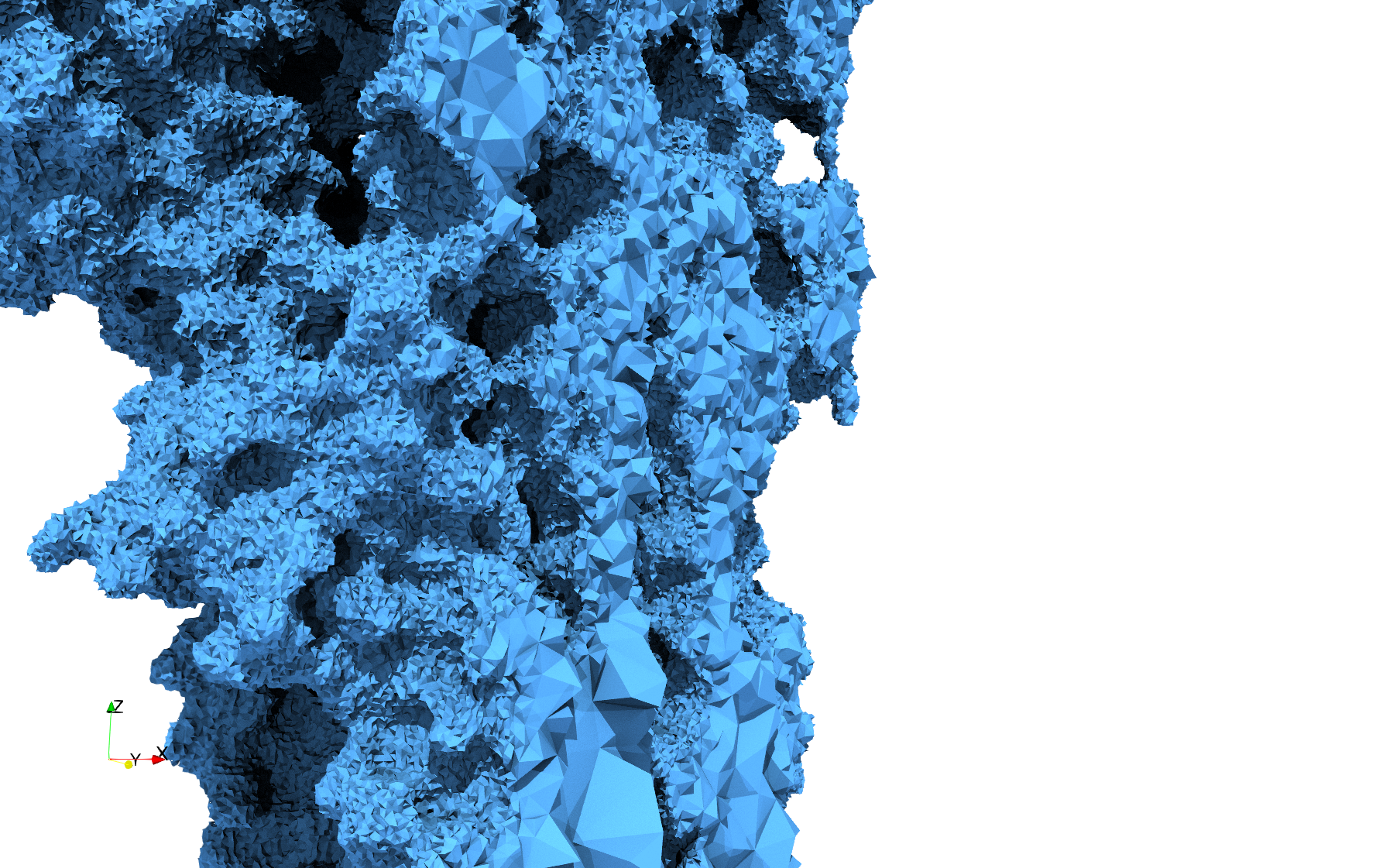
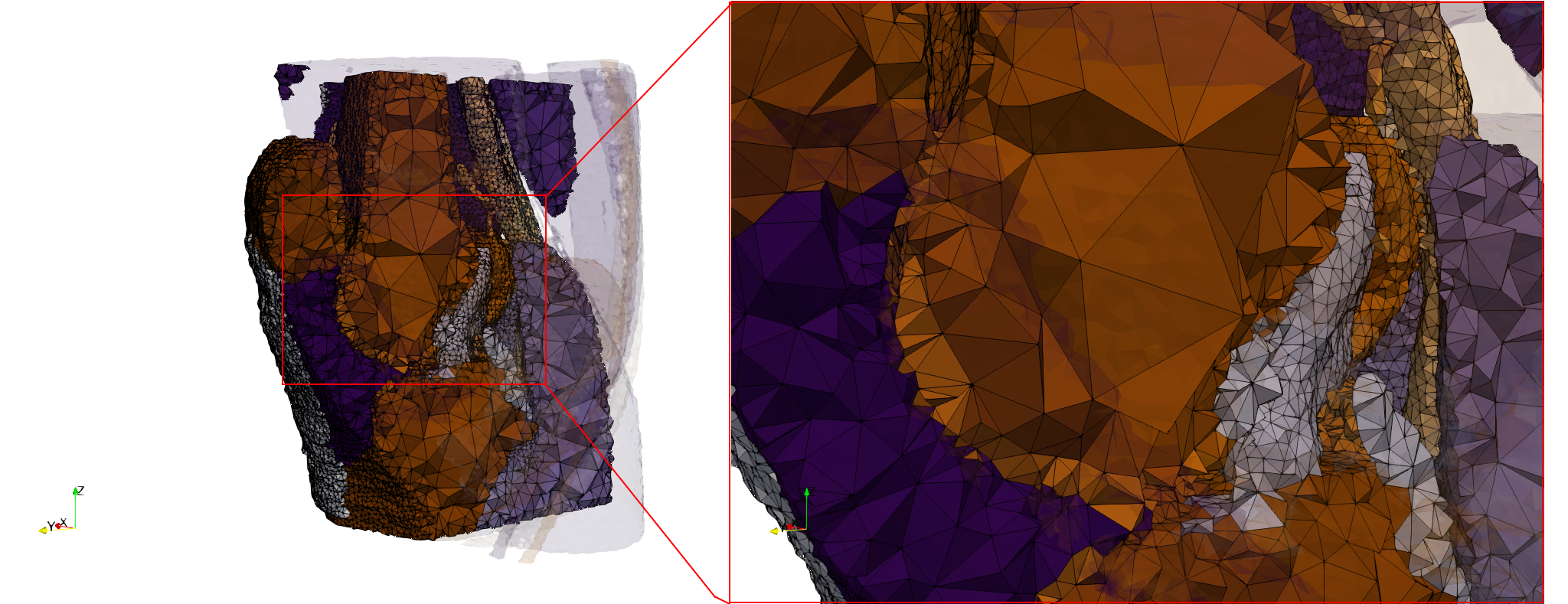
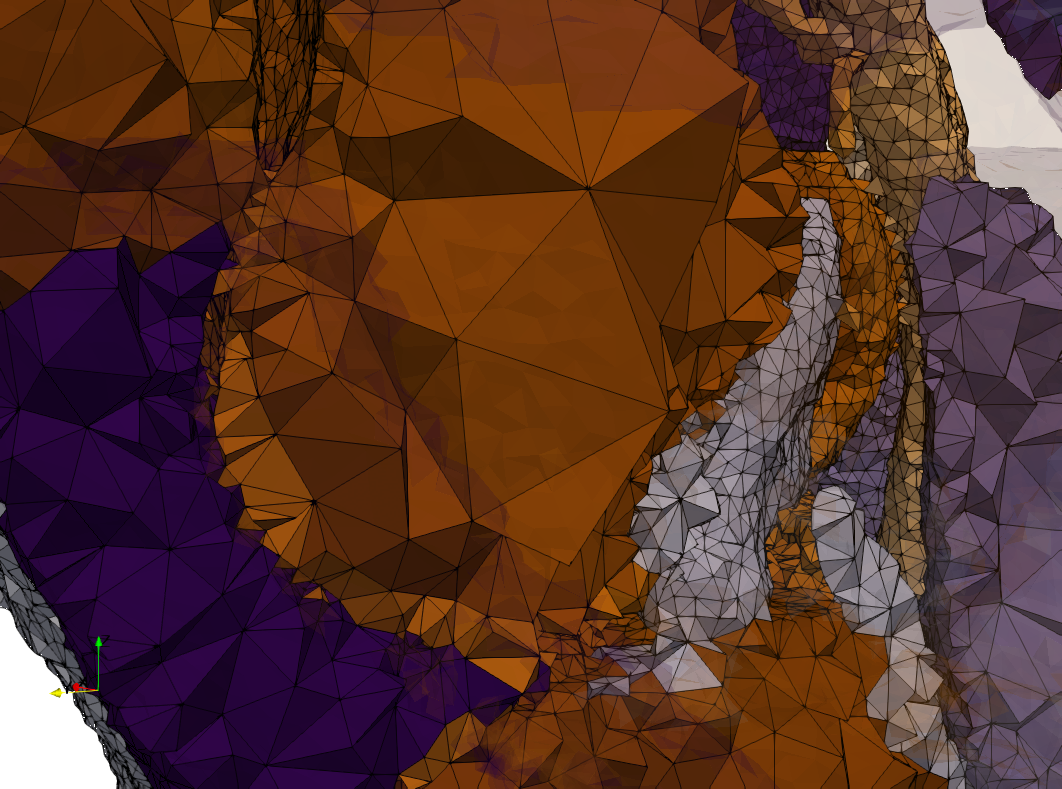
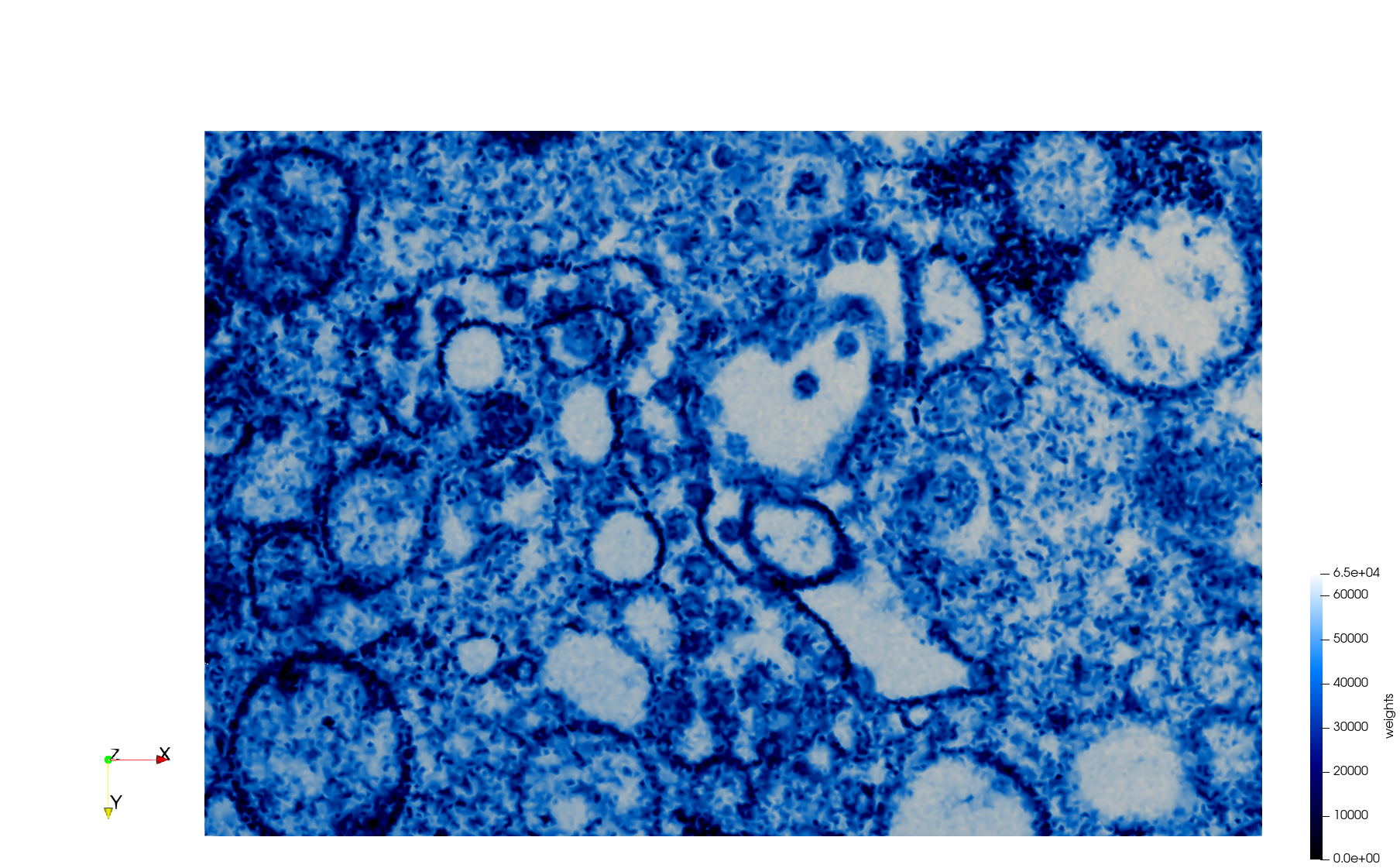
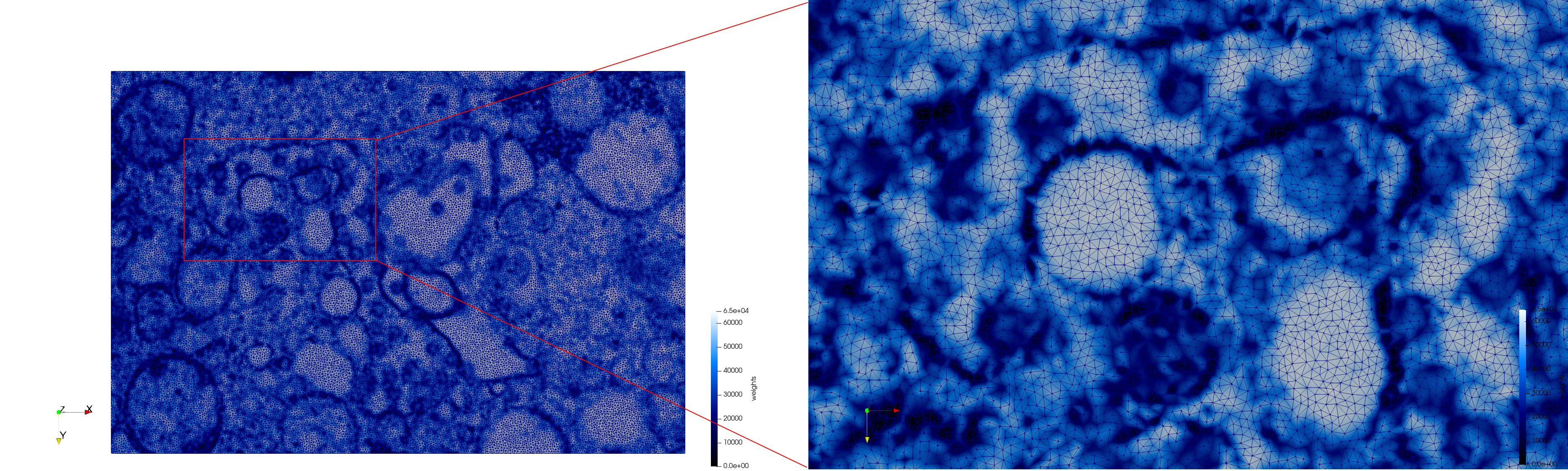
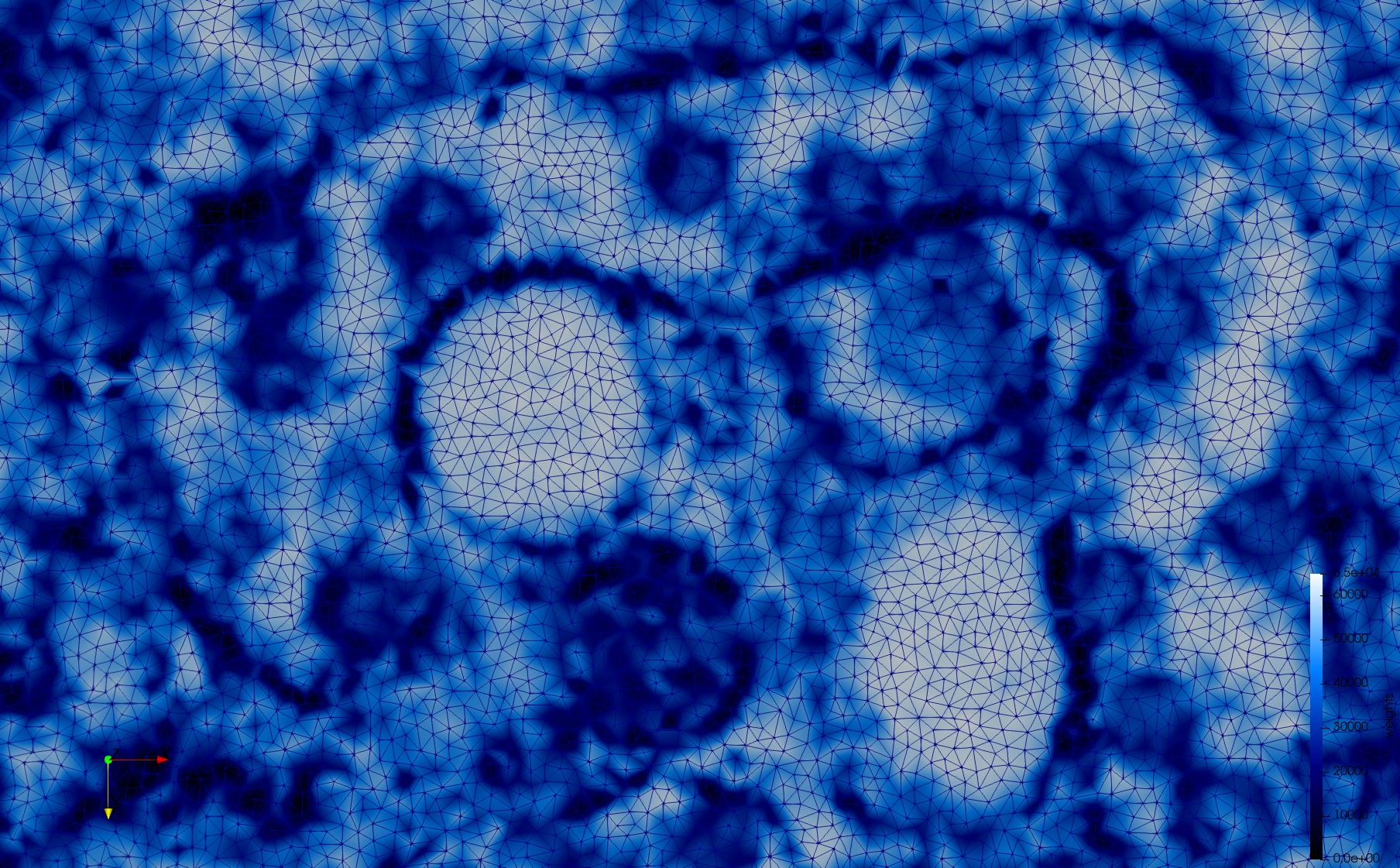
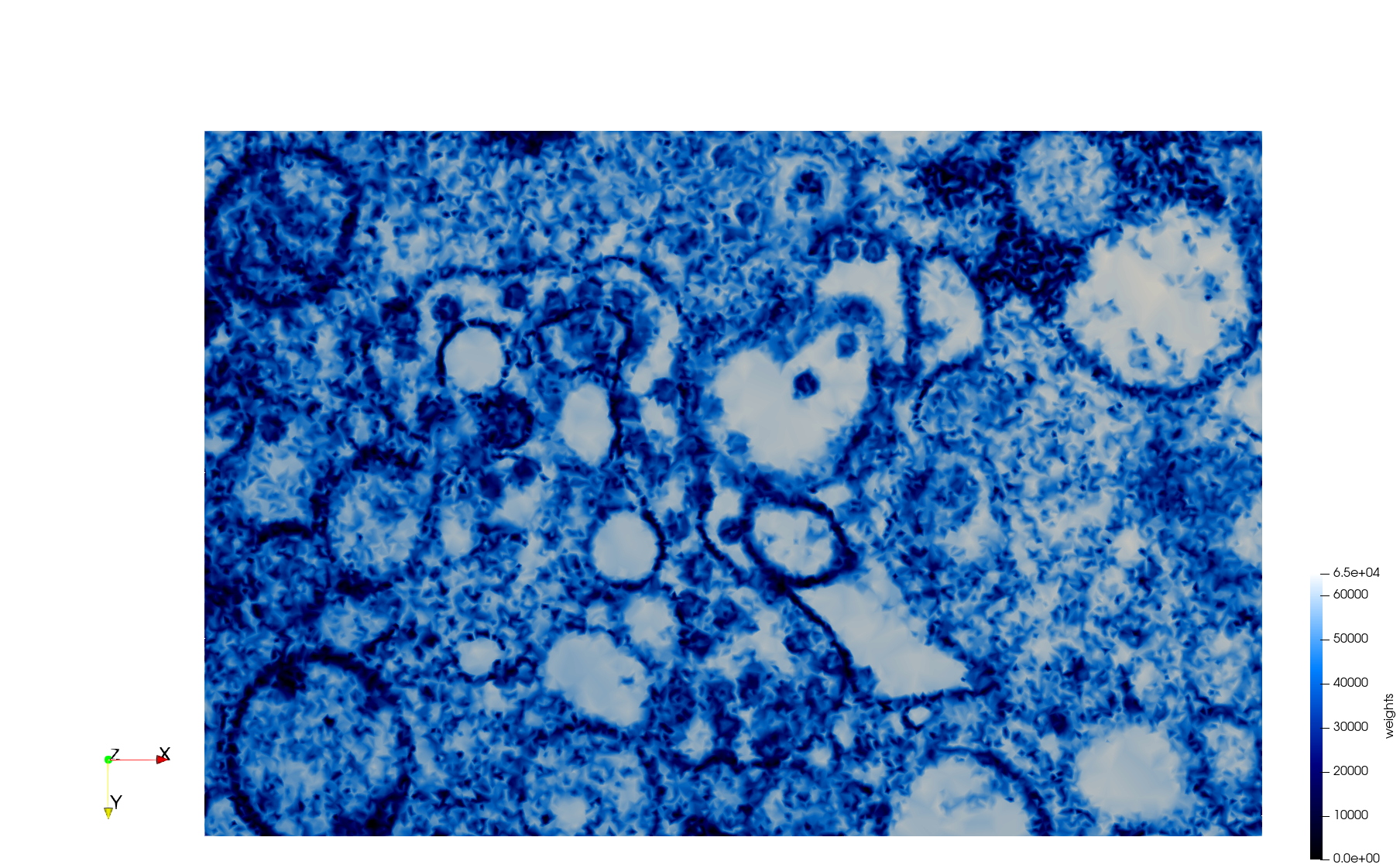
COVID-19-Main-Protease-6y2e
- Input image : Dimensions () with spacing ()
- Uniform with Delta = 0.5: 2,509,202 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Main-Protease-6y2e.nrrd --delta 0.5 --output ./COVID-19-Main-Protease-6y2e,d=0.5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 Main Protease. Biological assembly data was retrieved from the molecule 6y2e, Protein Data Bank.
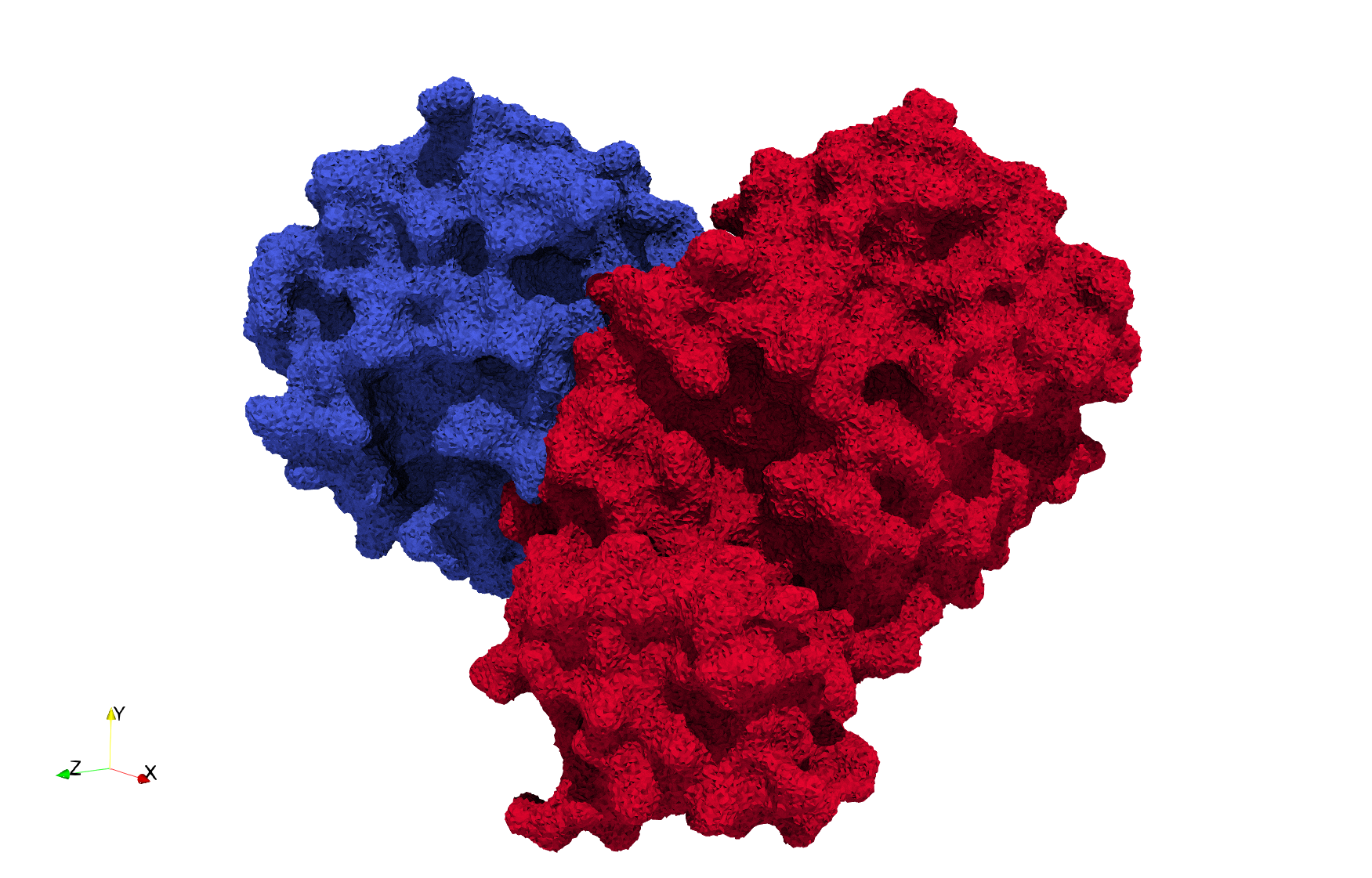
COVID-19-Spike-Glycoprotein-6vsb
- Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042)
- Uniform with Delta = 0.4: 5,731,833 tetrahedra
- Graded with Delta = 0.4: 3,794,223 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4.vtk
Graded with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --volume-grading --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.vtk
Uniform and graded tetrahedral meshes capable to help identify the internal voids of the COVID-19 spike glycoprotein. The middle and right meshes are slices. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
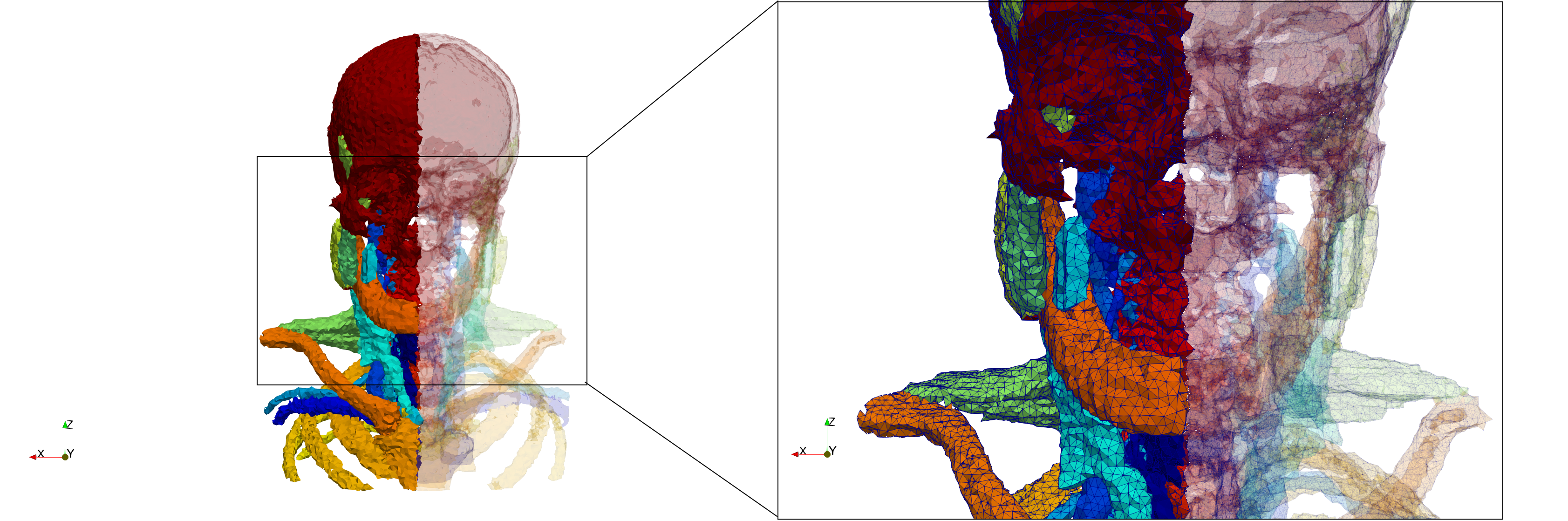
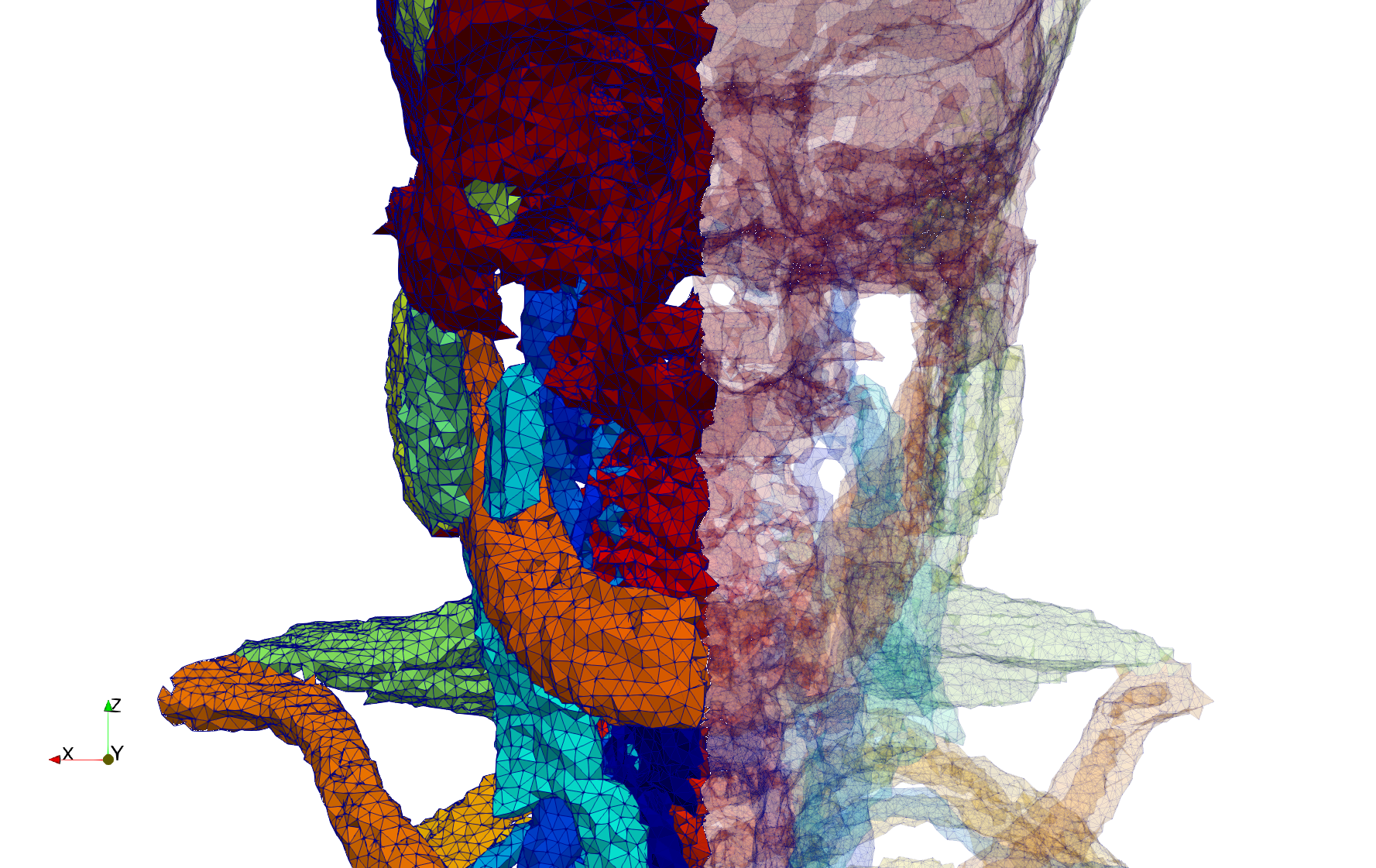
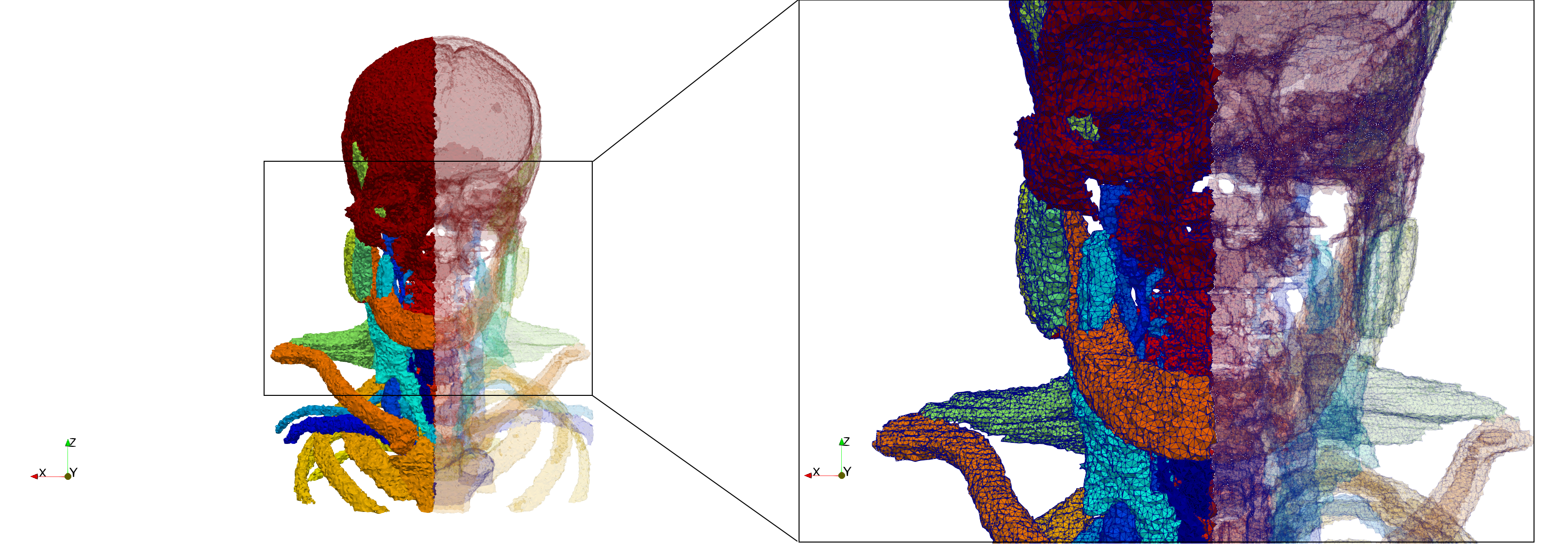
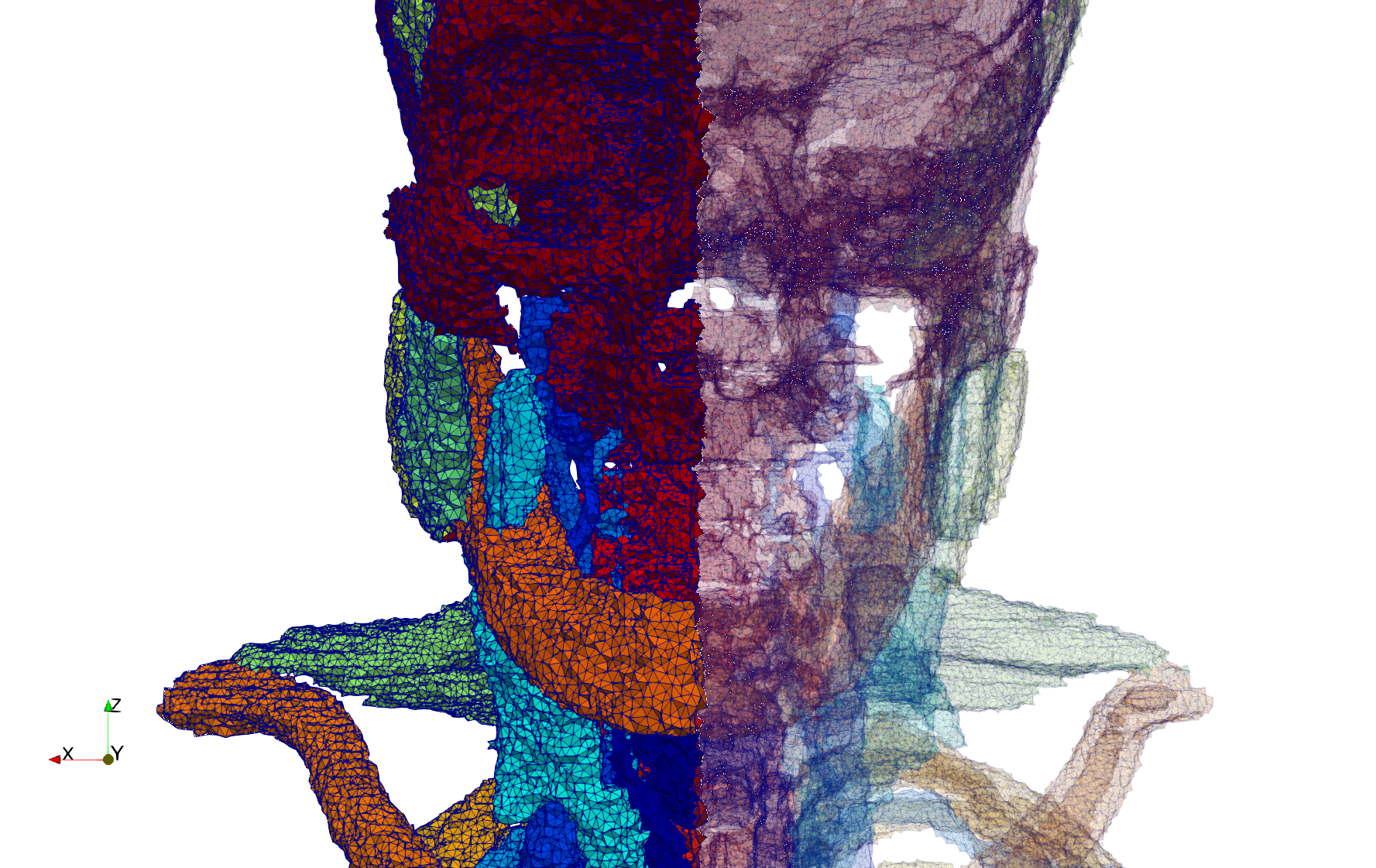
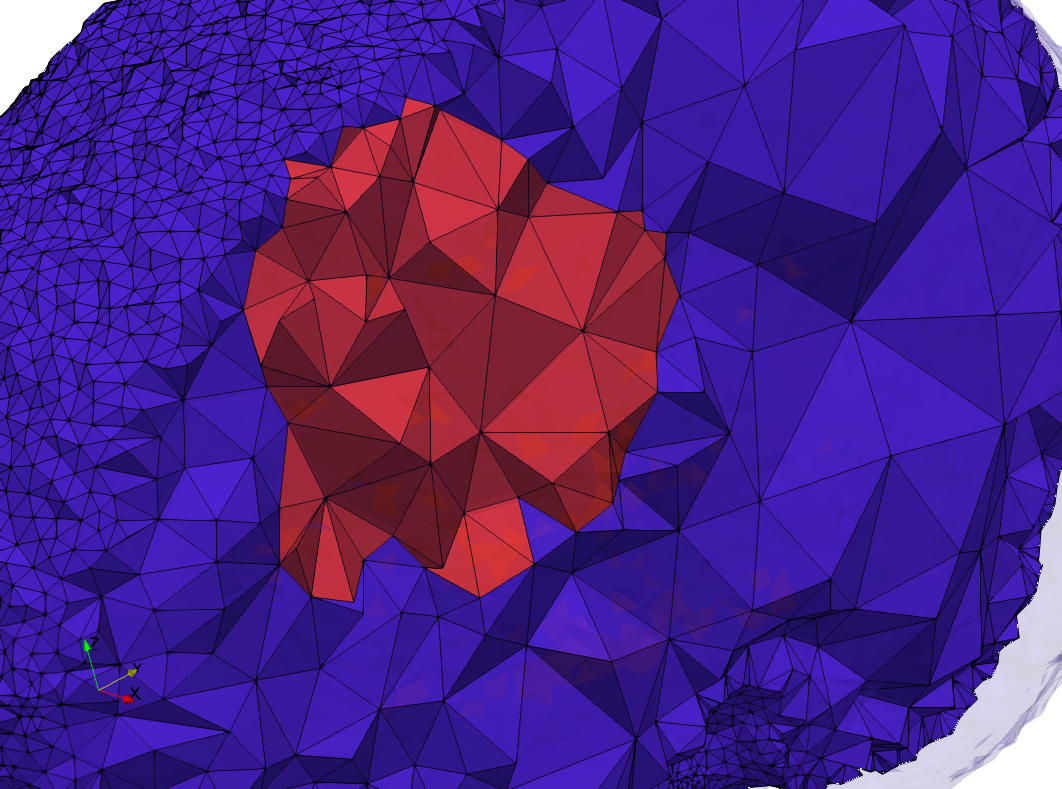
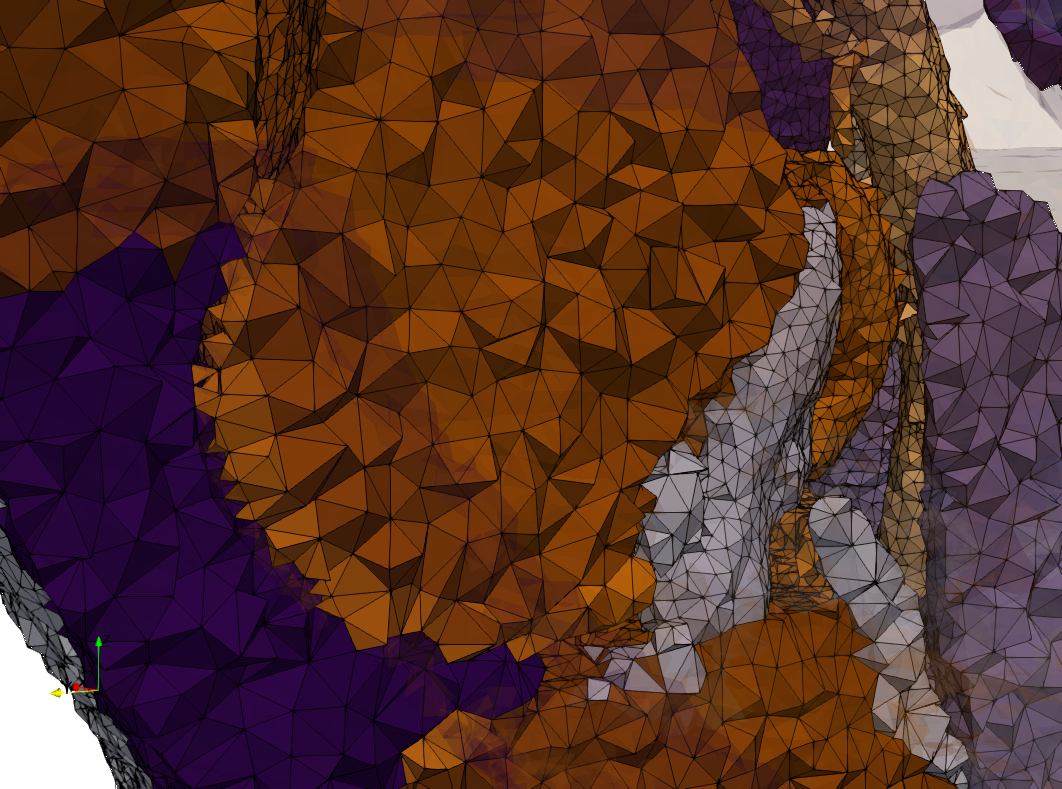
Head-Neck
- Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = default: 205,510 tetrahedra
- Uniform with Delta = 1.5: 767,393 tetrahedra
Commands to generate meshes:
Uniform with Delta = default: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --output ./Head-Neck,d=2.49023.vtk
Uniform with Delta = 1.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
Brain-With-Tumor-Case17
- Input image : Dimensions (448x512x176) with spacing (0.488281x0.488281x1)
- Uniform with Delta = default: 222,540 tetrahedra
- Graded with Delta = default: 94,383 tetrahedra
Commands to generate meshes:
Uniform with Delta = default: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --output ./Brain-With-Tumor-Case17,d=1.76001.vtk
Graded with Delta = default: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --volume-grading --output ./Brain-With-Tumor-Case17,d=1.76001,graded.vtk
Knee-Char
- Input image : Dimensions (512x512x119) with spacing (0.27734x0.27734x1)
- Uniform with Delta = default : 386,869 tetrahedra
- Graded with Delta = default : 274,309 tetrahedra
Commands to generate meshes:
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Knee-Char.mha --output ./Knee-Char,d=1.19.vtk
Graded with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Knee-Char.mha --volume-grading --output ./Knee-Char,d=1.19,graded.vtk
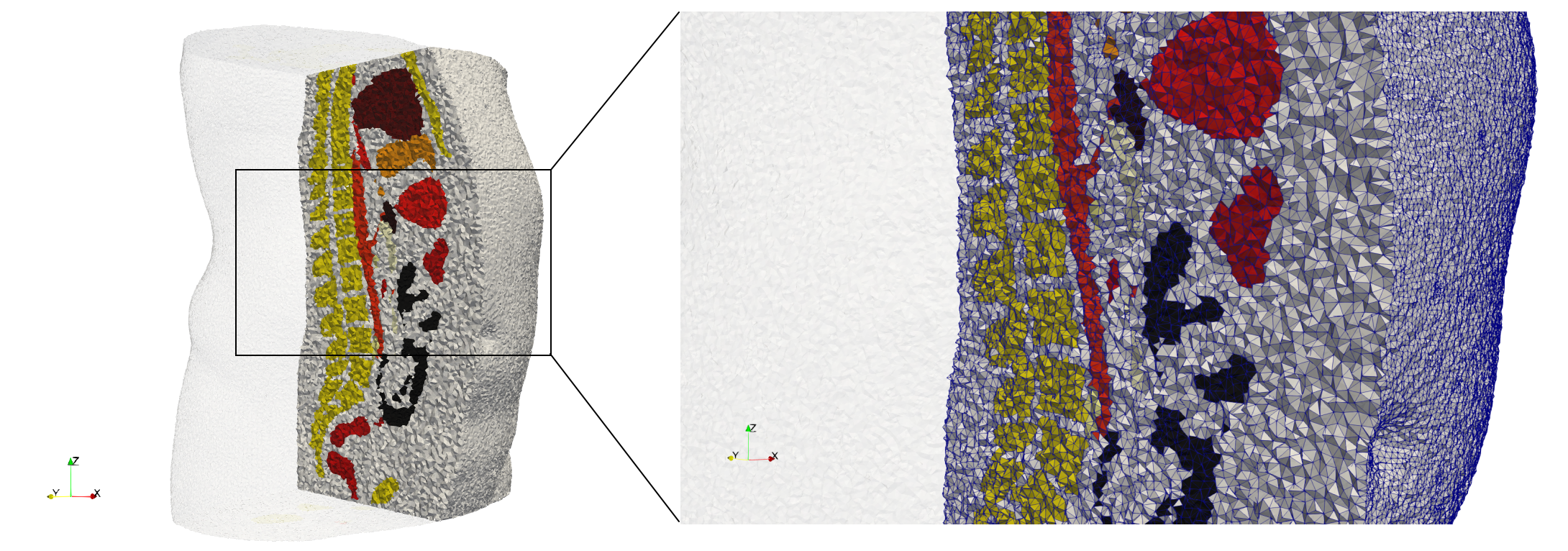
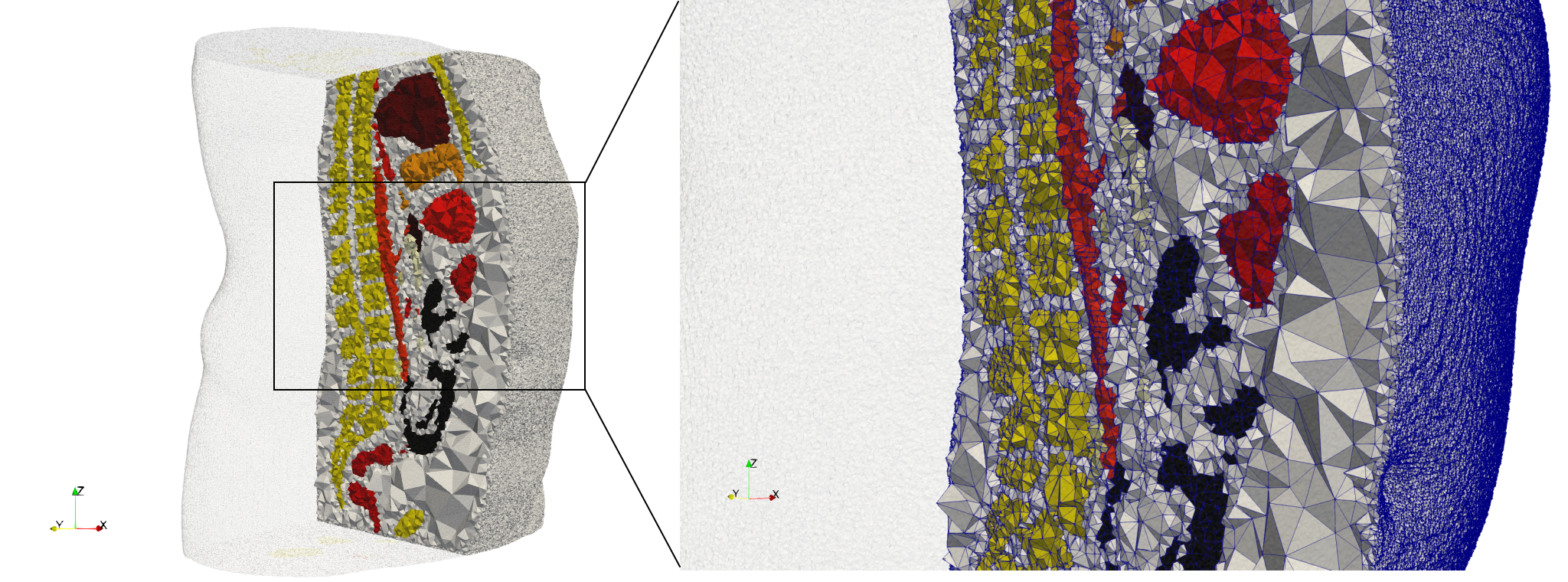
Ircad2
- Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = 2: 5,031,442 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
- Graded with Delta = 1: 6,072,751 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
Commands to generate meshes:
Uniform with Delta = 2: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 2 --threads 6 --output ./Ircad2,d=2.vtk
Graded with Delta = 1: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 6 --volume-grading --output ./Ircad2,d=1,graded.vtk
2D Example Meshes
The directory containing the 2D input data is located in the 2D folder of Medical_Imaging_Data.
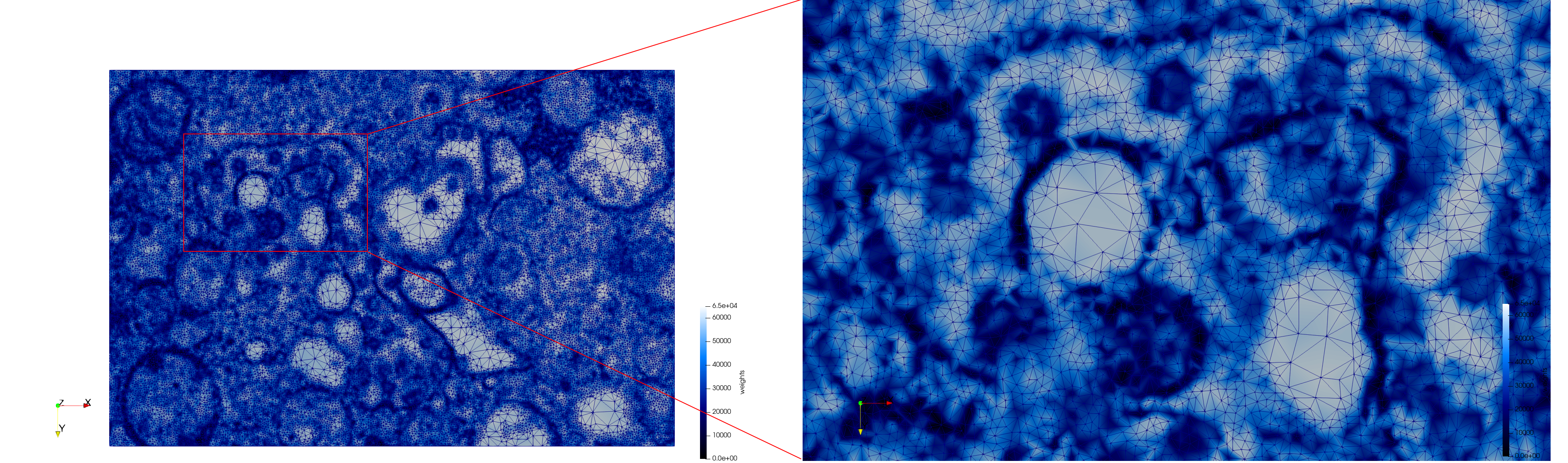
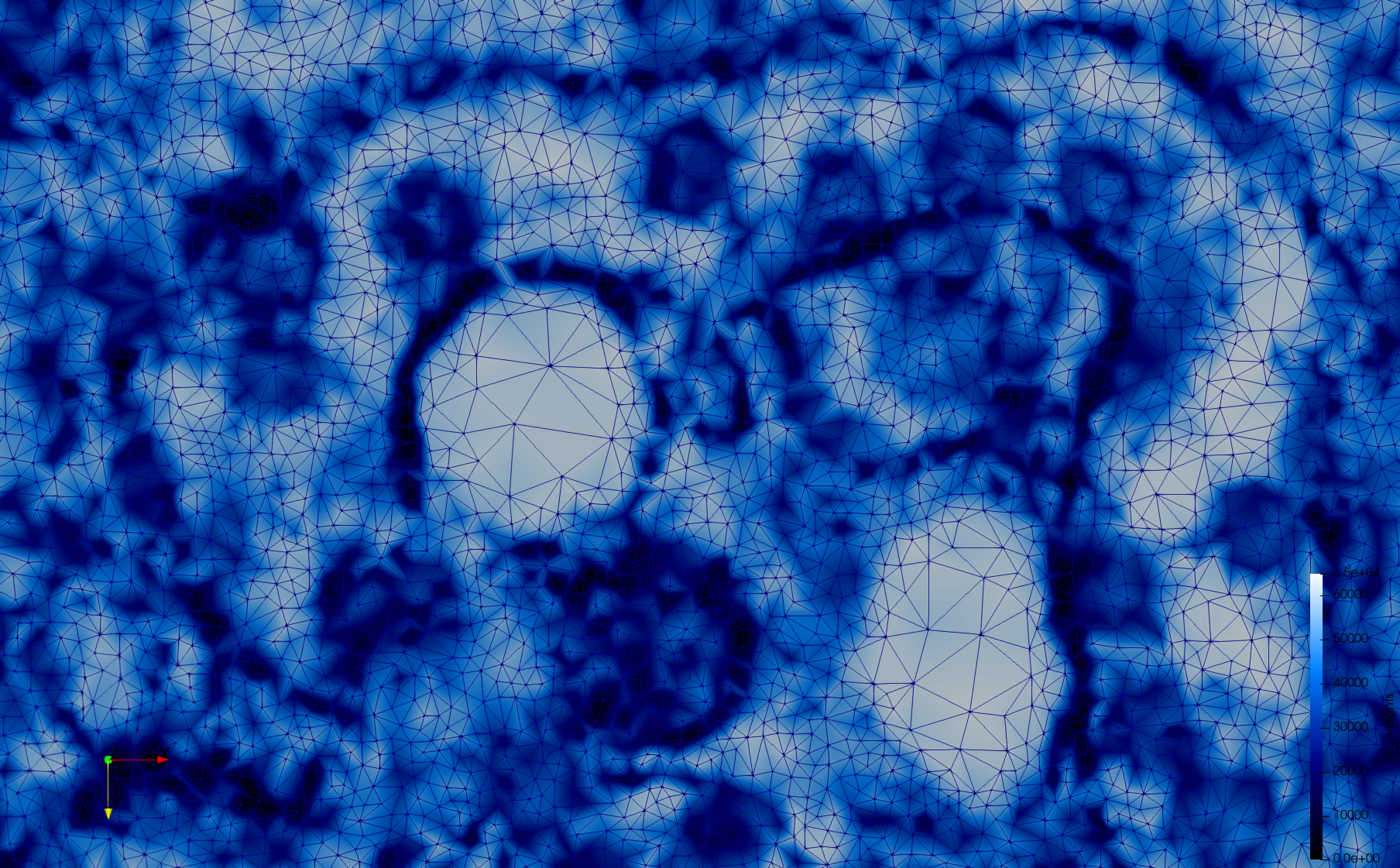
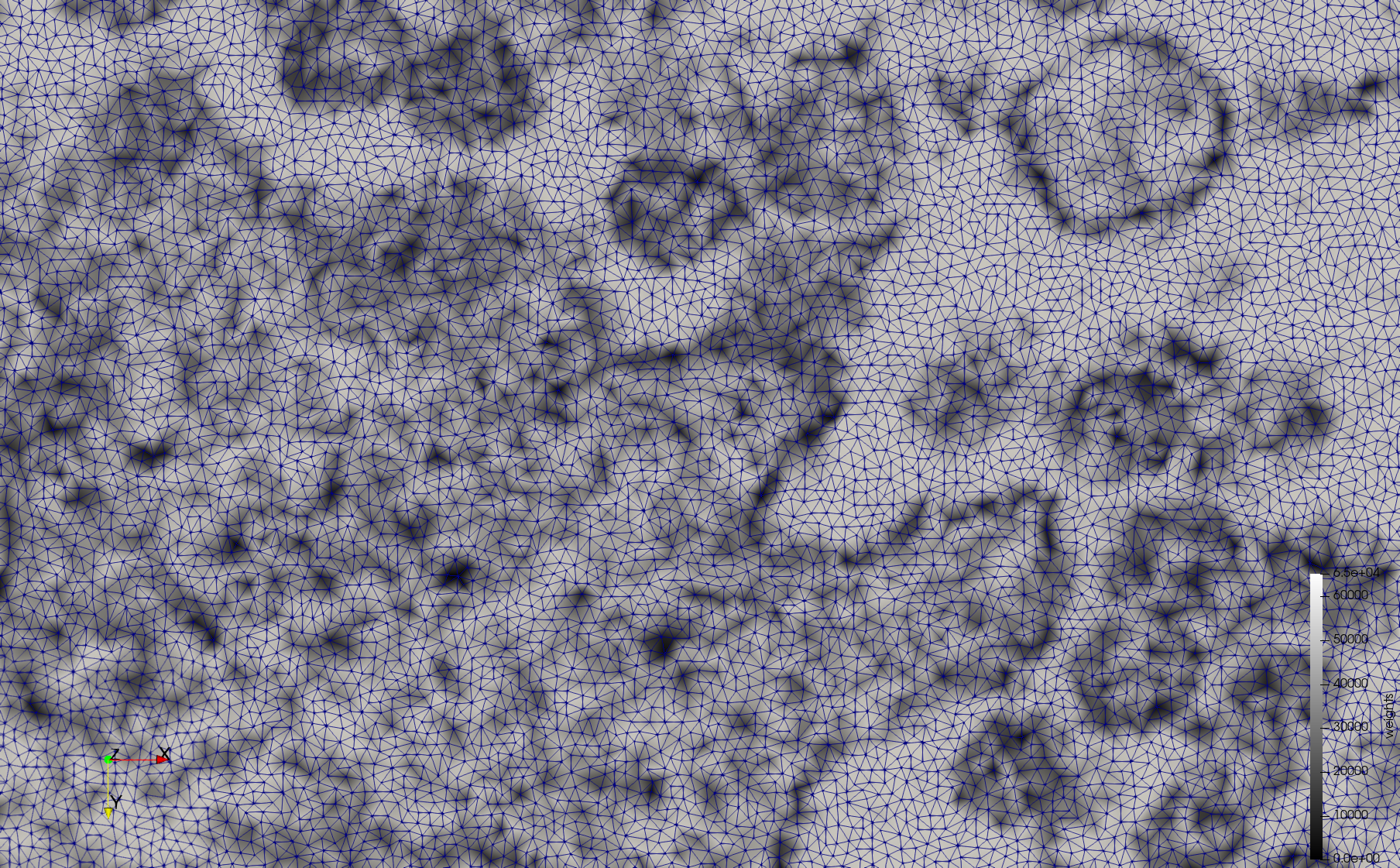
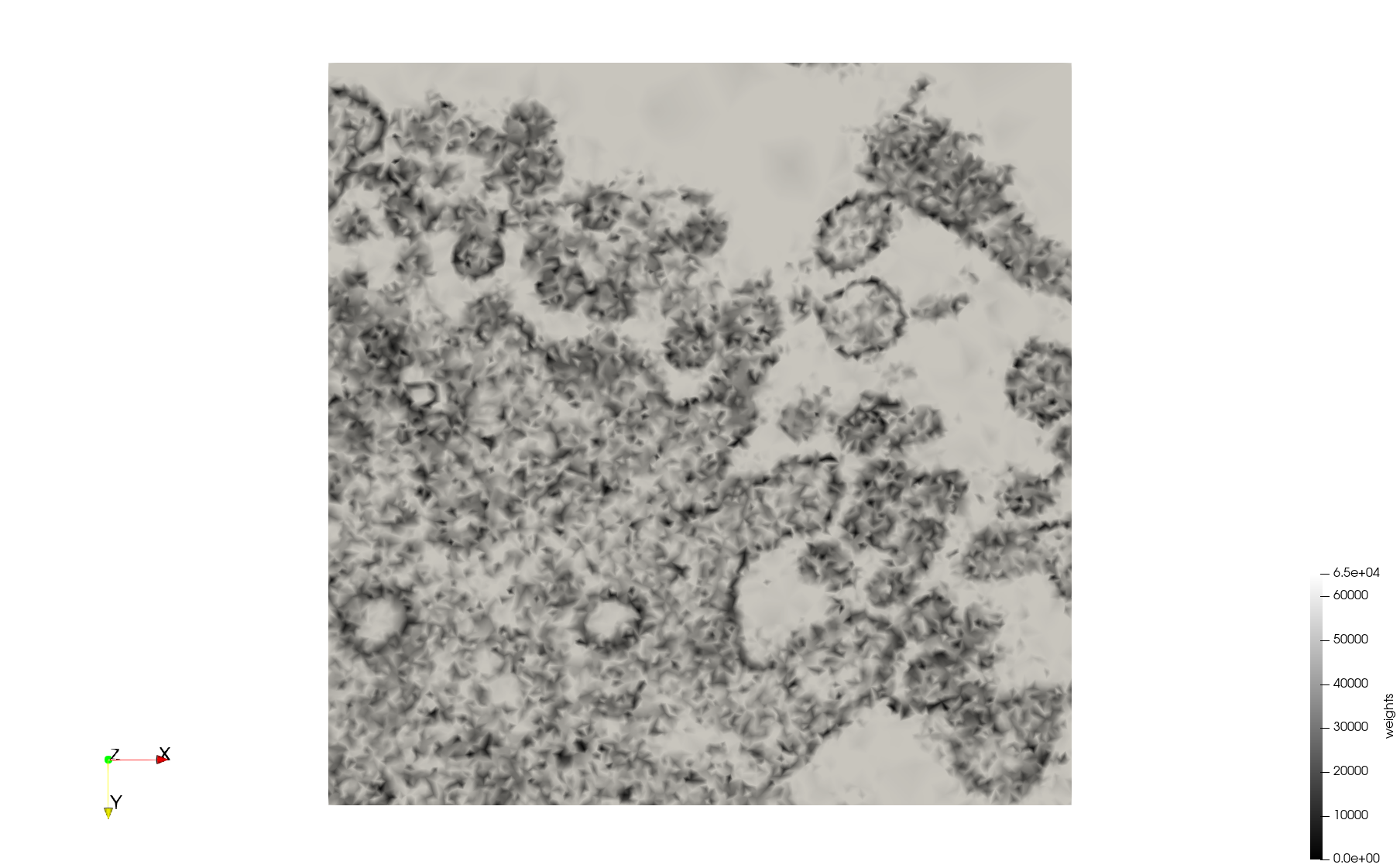
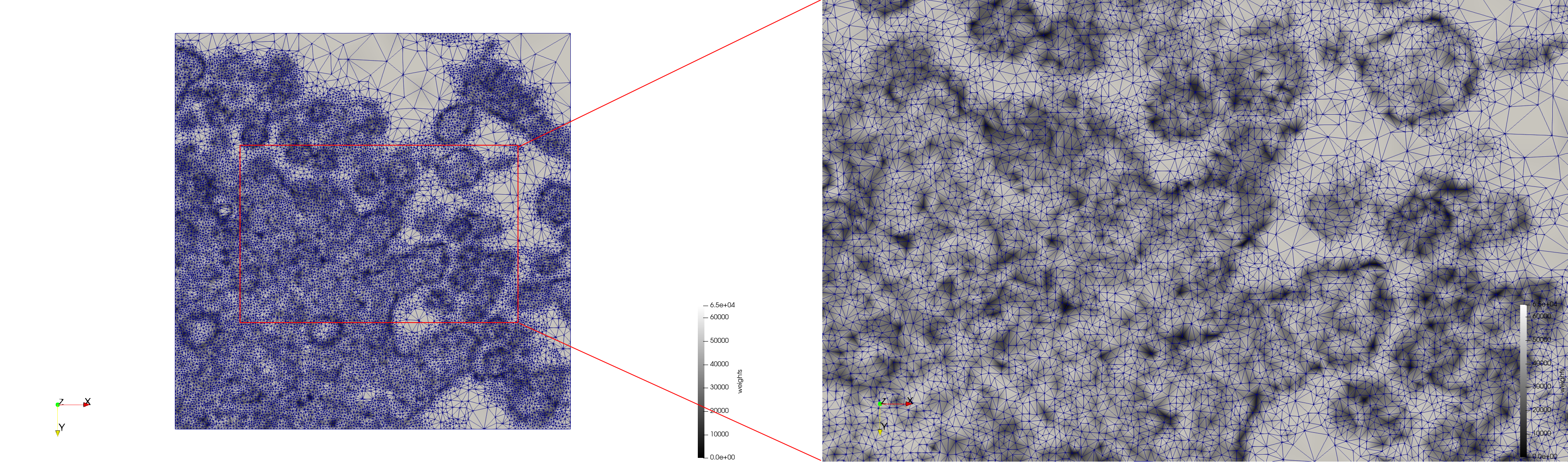
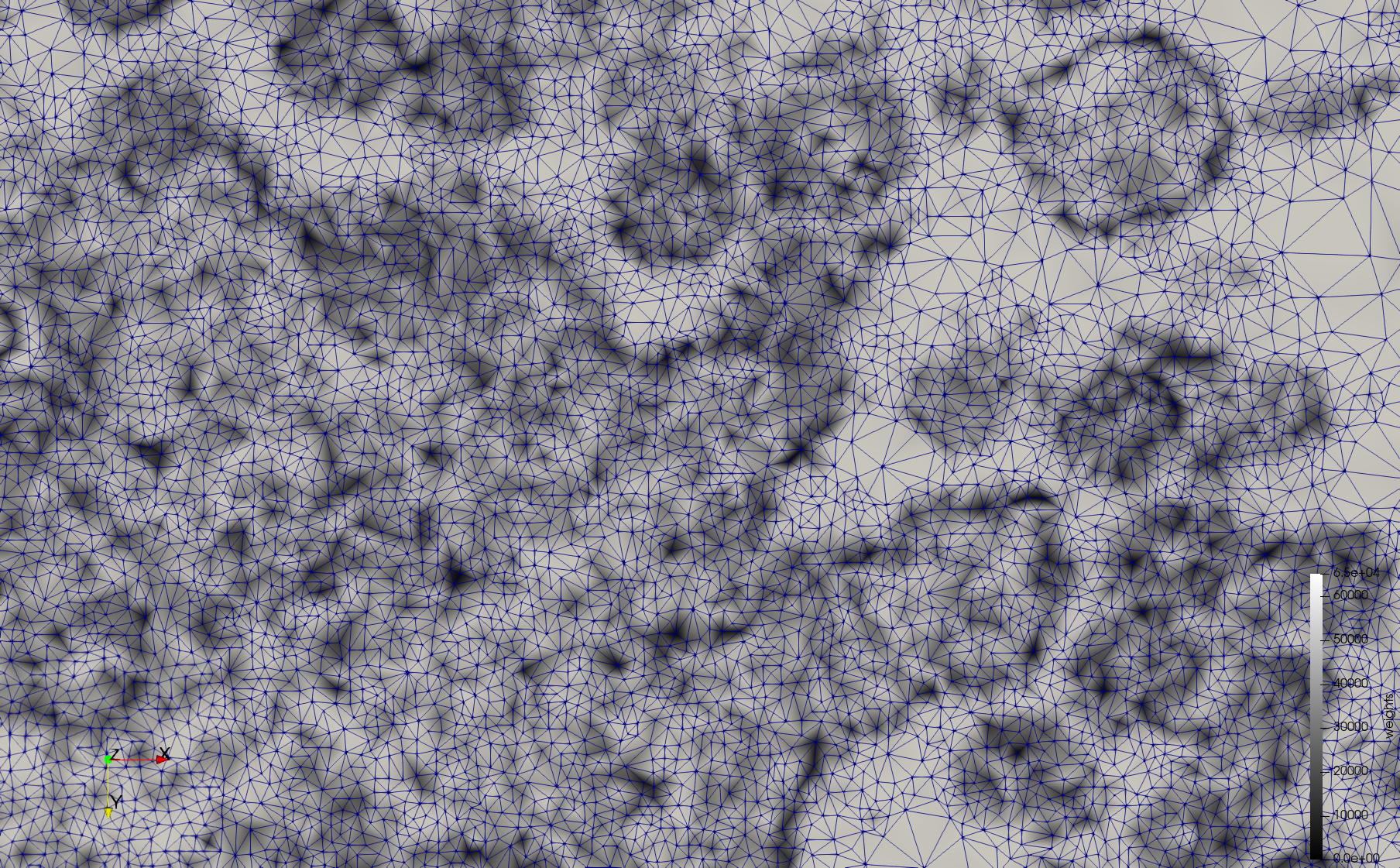
COVID-19-23354
- Input image : Dimensions (3,000x2,000) with spacing (1x1)
- Uniform with Min-Edge = 15: 82,981 triangles
- Adaptive with Min-Edge = 15: 67,920 triangles
Commands to generate meshes:
Uniform with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --uniform --min-edge 15 --output ./COVID-19-23354,uniform,e=15.vtk
Adaptive with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --min-edge 15 --output ./COVID-19-23354,w=0.1,e=15.vtk
Mesh generated based on the image 23354 retrieved from the Centers for Disease Control and Prevention. For the uniform meshes, the edge-size corresponds to 15 pixels. The adaptive meshes were created by controlling the size of the elements based on the difference in the intensity of the pixels.
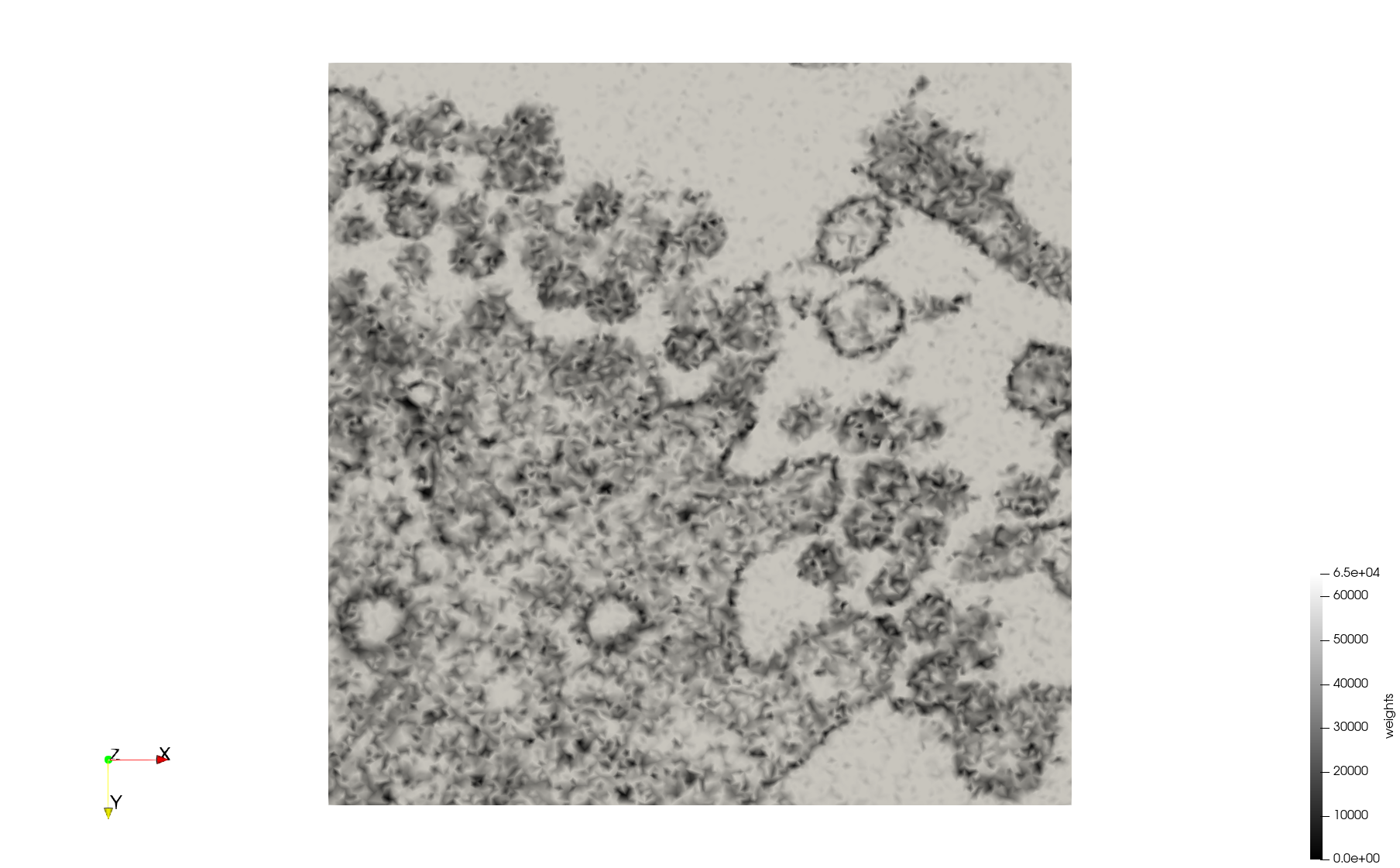
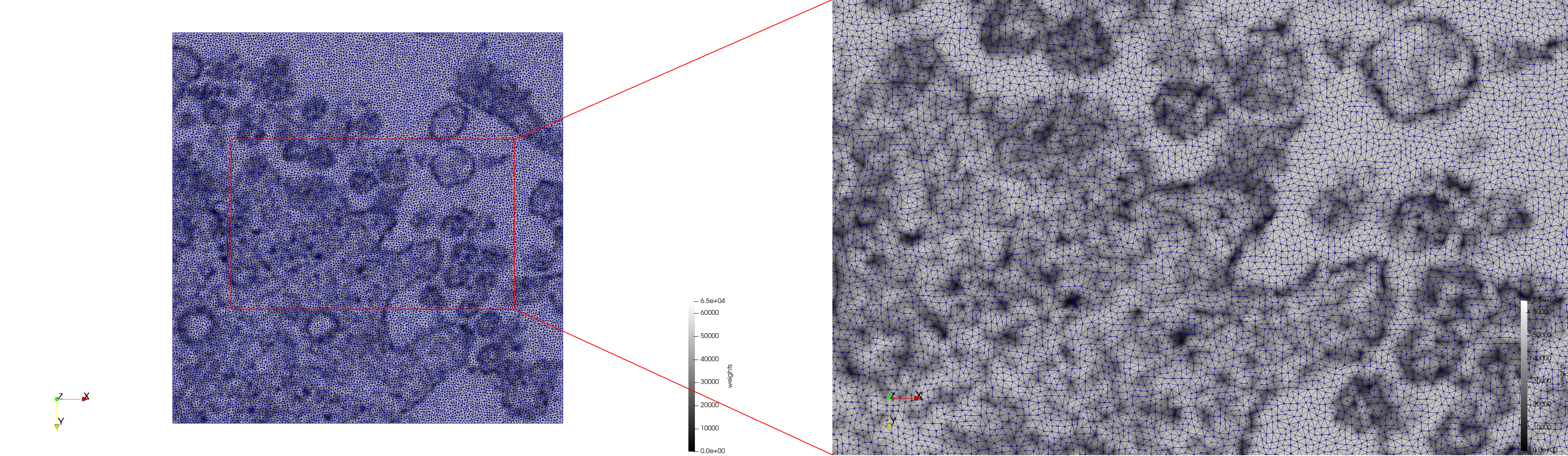
COVID-19-23311
- Input image : Dimensions (2,460x2,460) with spacing (1x1)
- Uniform with Min-Edge = 20: 47,028 triangles
- Adaptive with Min-Edge = 20: 34,080 tetrahedra
Commands to generate meshes:
Uniform with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --uniform --min-edge 20 --output ./COVID-19-23311,uniform,e=20.vtk
Adaptive with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --min-edge 20 --output ./COVID-19-23311,w=0.1,e=20.vtk
Mesh generated based on the image 23311 retrieved from the Centers for Disease Control and Prevention. For the uniform meshes, the edge-size corresponds to 20 pixels. The adaptive meshes were created by controlling the size of the elements based on the difference in the intensity of the pixels.