Difference between revisions of "Medical Imaging Example Meshes"
Spyridon97 (talk | contribs) (→Head-Neck) |
Spyridon97 (talk | contribs) (→3D Example Meshes) |
||
| Line 168: | Line 168: | ||
<pre> | <pre> | ||
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 6 --volume-grading --output ./Ircad2,d=1,graded.vtk | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 6 --volume-grading --output ./Ircad2,d=1,graded.vtk | ||
| + | </pre> | ||
| + | |||
| + | ==COVID-19-Spike-Glycoprotein-6vsb== | ||
| + | * Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042) | ||
| + | * Uniform with Delta = 0.5: 5,731,833 tetrahedra | ||
| + | * Graded with Delta = 0.4: 3,794,223 tetrahedra | ||
| + | |||
| + | [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4.png] | ||
| + | [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip.png] | ||
| + | [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip,zoom.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip,zoom.png] | ||
| + | <gallery mode="packed" heights=350px> | ||
| + | </gallery> | ||
| + | [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.png] | ||
| + | [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip.png] | ||
| + | [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip,zoom.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip,zoom.png] | ||
| + | <gallery mode="packed" heights=350px> | ||
| + | </gallery> | ||
| + | |||
| + | Commands to generate meshes : | ||
| + | |||
| + | '''Uniform with Delta = 0.4:''' [ Output Mesh] | ||
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4.vtk | ||
| + | </pre> | ||
| + | |||
| + | '''Graded with Delta = 0.4:''' [https://odu.box.com/s/2kn1ovwpa6jarj8wssmeqz2y5zc19fc2 Output Mesh] | ||
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --volume-grading --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.vtk | ||
</pre> | </pre> | ||
Revision as of 00:58, 16 March 2020
Contents
2D Example Meshes
The directory containing the 2D input data is located in the 2D folder of Medical_Imaging_Data.
COVID-19-23354
- Input image : Dimensions (3,000x2,000) with spacing (1x1)
- Uniform with Min-Edge = 15: 82,981 triangles
- Adaptive with Min-Edge = 15: 67,920 triangles
Commands to generate meshes :
Uniform with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --uniform --min-edge 15 --output ./COVID-19-23354,uniform,e=15.vtk
Adaptive with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --min-edge 15 --output ./COVID-19-23354,w=0.1,e=15.vtk
Mesh generated based on the images published at Centers for Disease Control and Prevention. For the uniform meshes, the edge-size corresponds to 15 pixels. The adaptive meshes were created by controlling the size of the elements based on the difference in the intensity of the pixels.
COVID-19-23311
- Input image : Dimensions (2,460x2,460) with spacing (1x1)
- Uniform with Min-Edge = 20: 47,028 triangles
- Adaptive with Min-Edge = 20: 34,080 tetrahedra
Commands to generate meshes :
Uniform with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --uniform --min-edge 20 --output ./COVID-19-23311,uniform,e=20.vtk
Adaptive with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --min-edge 20 --output ./COVID-19-23311,w=0.1,e=20.vtk
Mesh generated based on the images published at Centers for Disease Control and Prevention. For the uniform meshes, the edge-size corresponds to 20 pixels. The adaptive meshes were created by controlling the size of the elements based on the difference in the intensity of the pixels.
3D Example Meshes
The directory containing the 3D input data is located in the 3D folder of Medical_Imaging_Data.
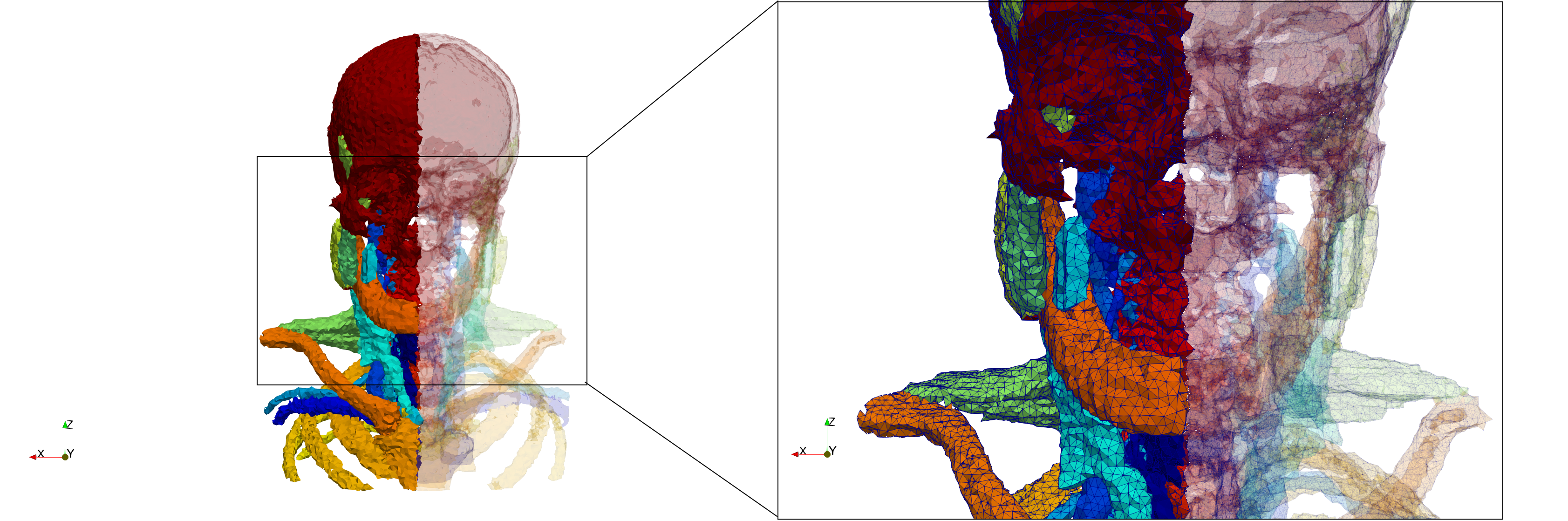
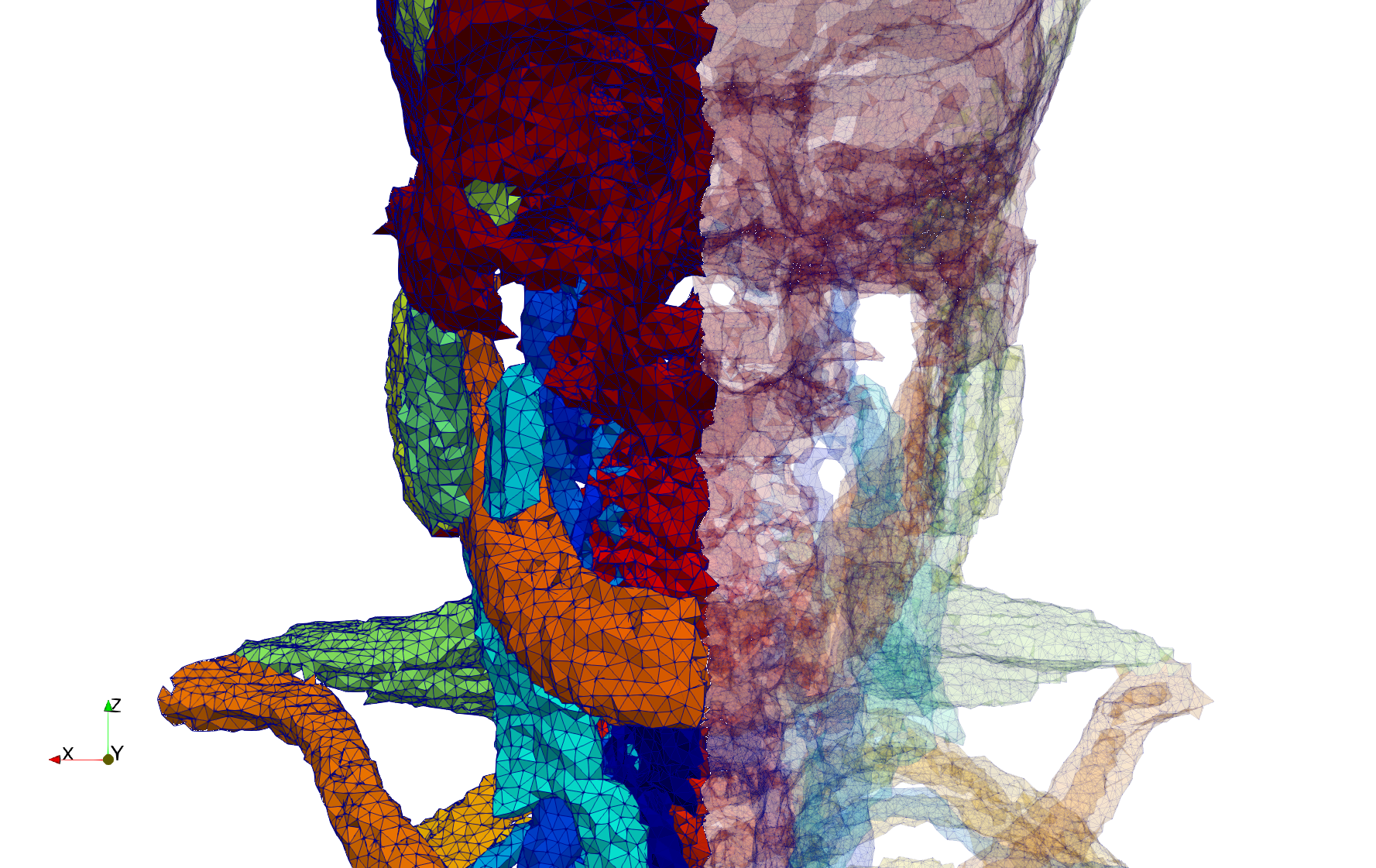
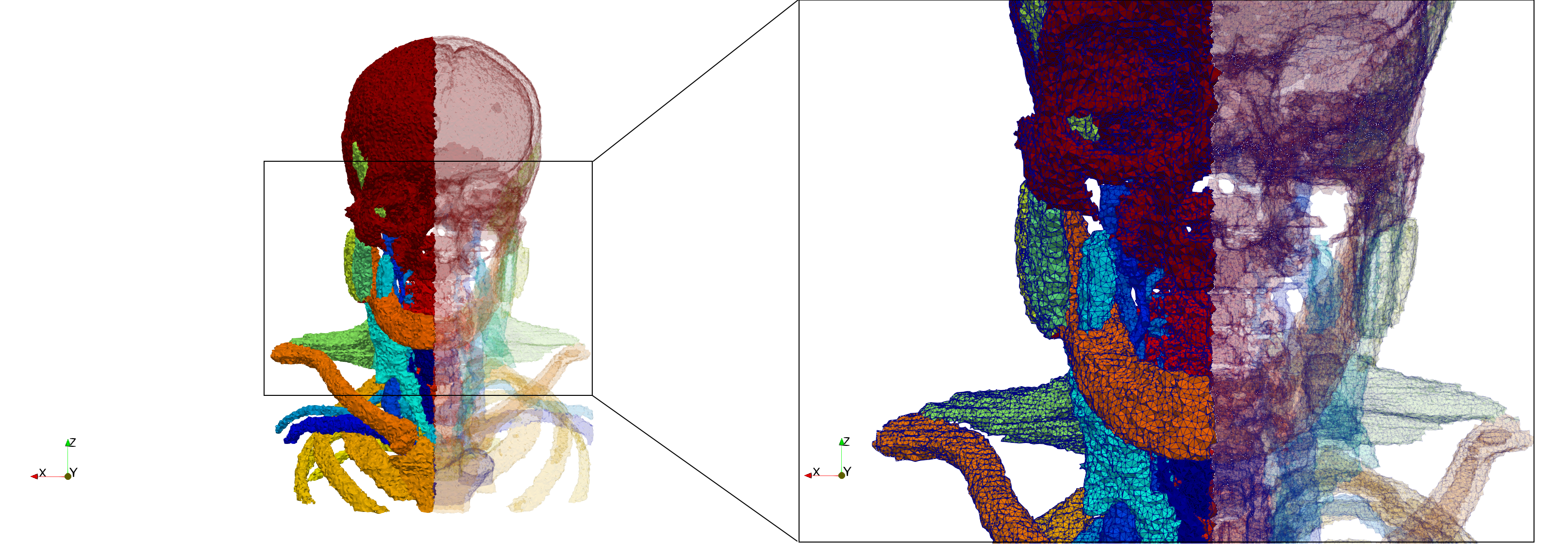
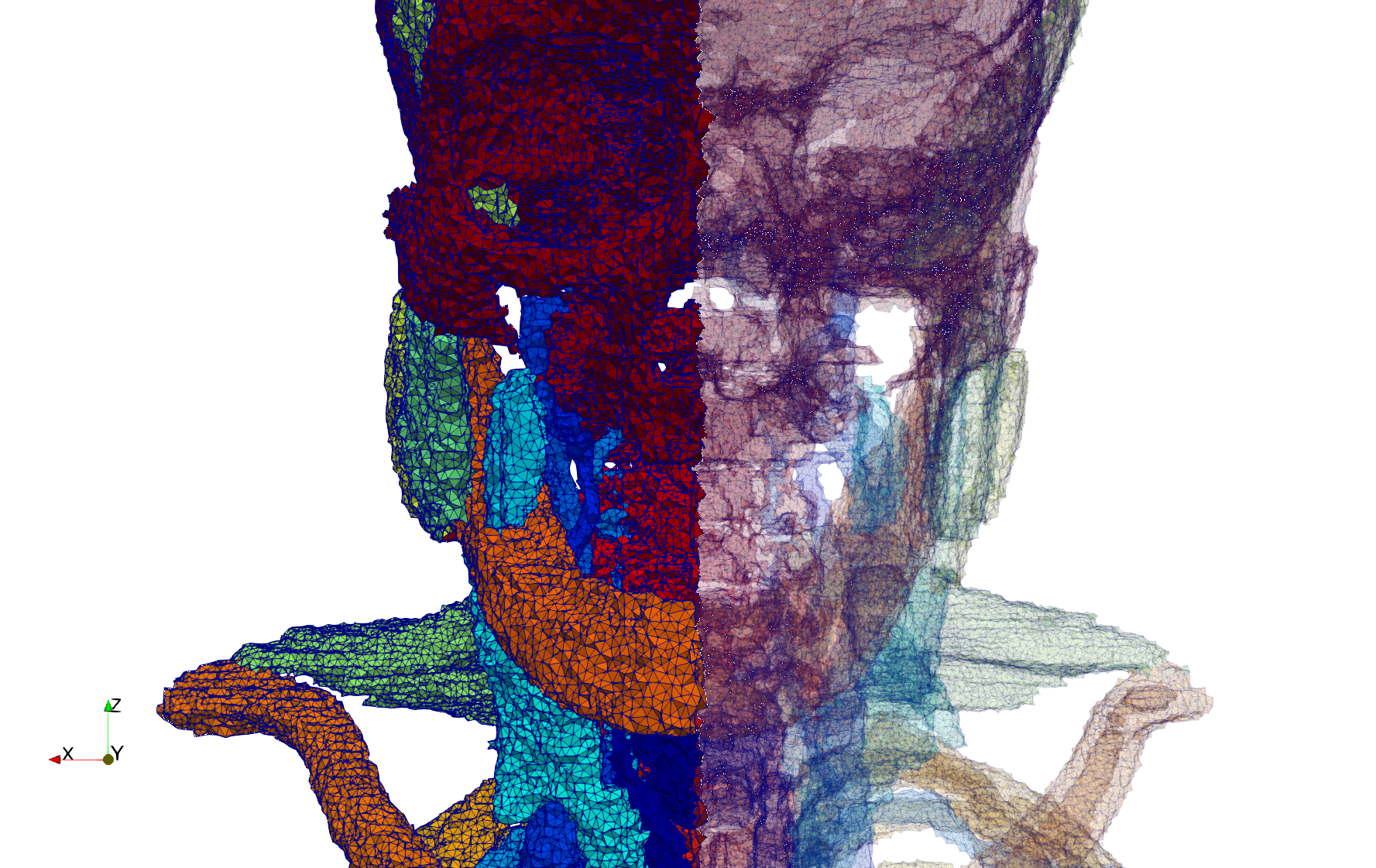
Head-Neck
- Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = default: 205,510 tetrahedra
- Uniform with Delta = 1.5: 767,393 tetrahedra
Commands to generate meshes :
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --output ./Head-Neck,d=2.49023.vtk
Uniform with Delta = 1.5 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
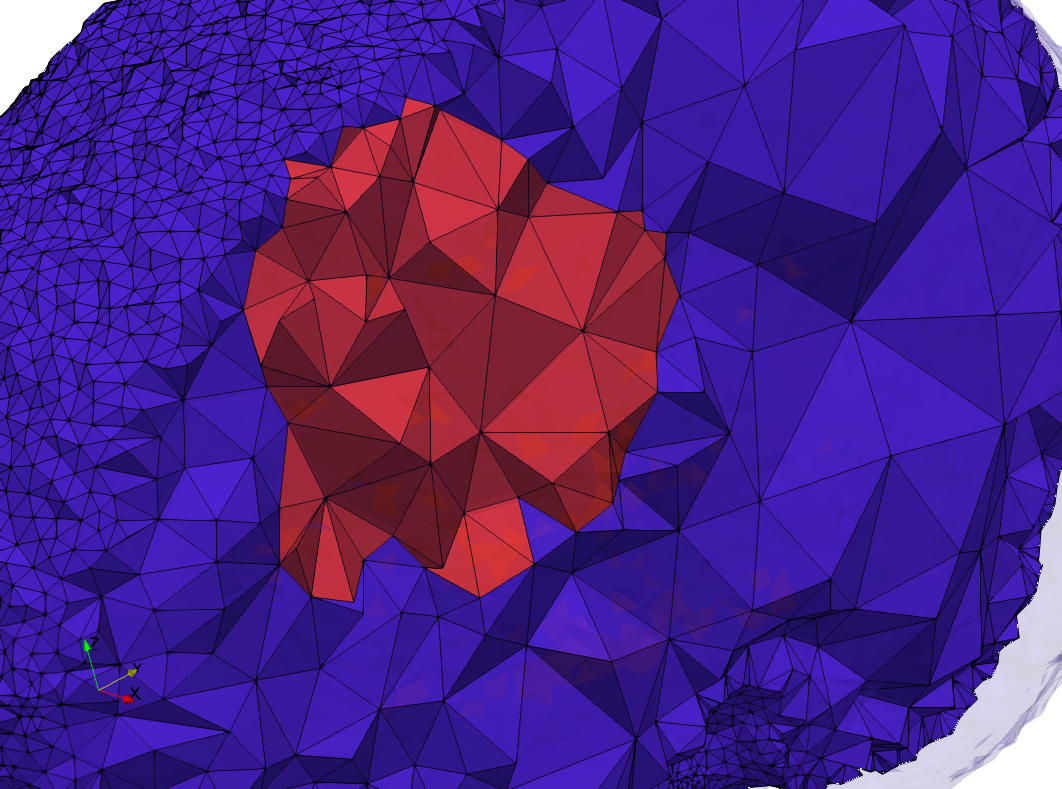
Brain-With-Tumor-Case17
- Input image : Dimensions (448x512x176) with spacing (0.488281x0.488281x1)
- Uniform with Delta = default: 222,540 tetrahedra
- Graded with Delta = default: 94,383 tetrahedra
Commands to generate meshes :
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --output ./Brain-With-Tumor-Case17,d=1.76001.vtk
Graded with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --volume-grading --output ./Brain-With-Tumor-Case17,d=1.76001,graded.vtk
Knee-Char
- Input image : Dimensions (512x512x119) with spacing (0.27734x0.27734x1)
- Uniform with Delta = default : 386,869 tetrahedra
- Graded with Delta = default : 274,309 tetrahedra
Commands to generate meshes :
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Knee-Char.mha --output ./Knee-Char,d=1.19.vtk
Graded with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Knee-Char.mha --volume-grading --output ./Knee-Char,d=1.19,graded.vtk
Ircad2
- Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = 2: 5,031,442 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
- Graded with Delta = 1: 6,072,751 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
Commands to generate meshes :
Uniform with Delta = 2 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 2 --threads 6 --output ./Ircad2,d=2.vtk
Graded with Delta = 1 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 6 --volume-grading --output ./Ircad2,d=1,graded.vtk
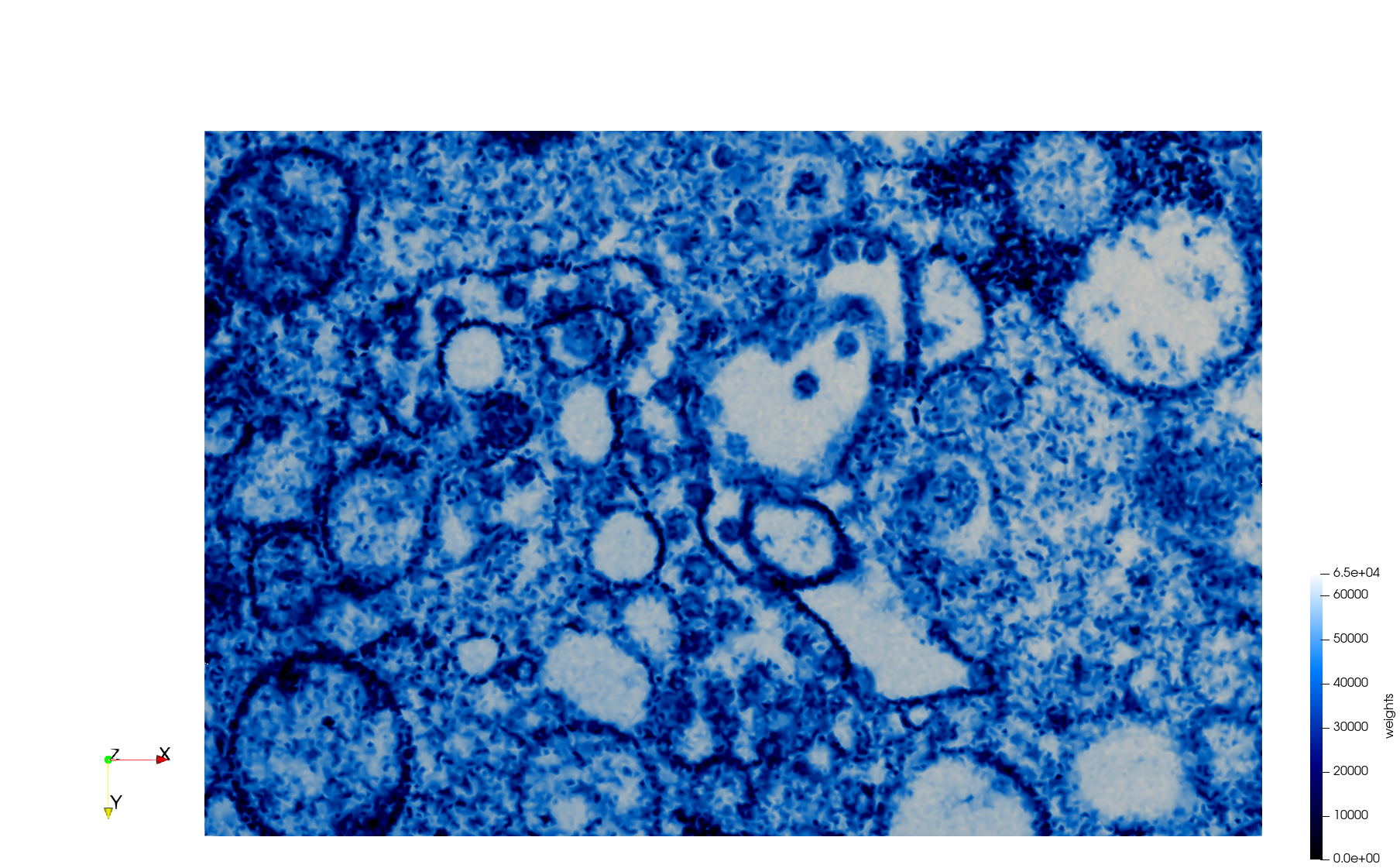
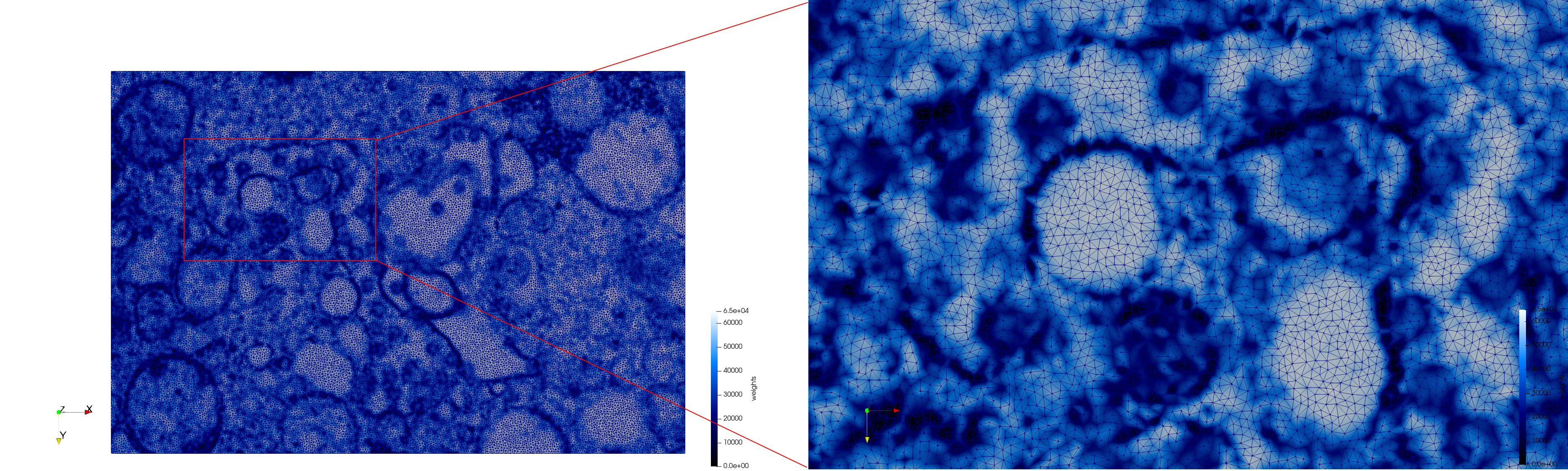
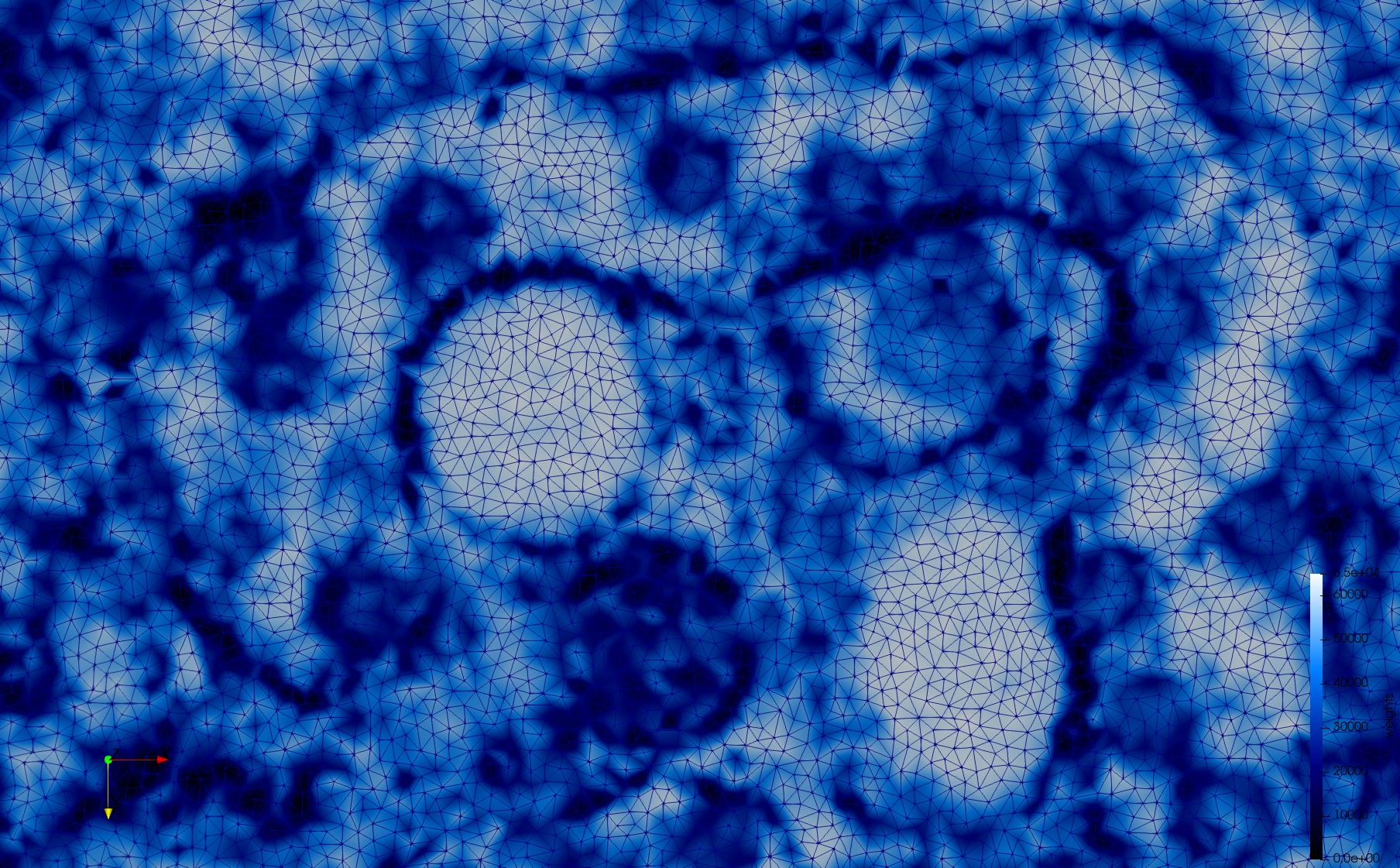
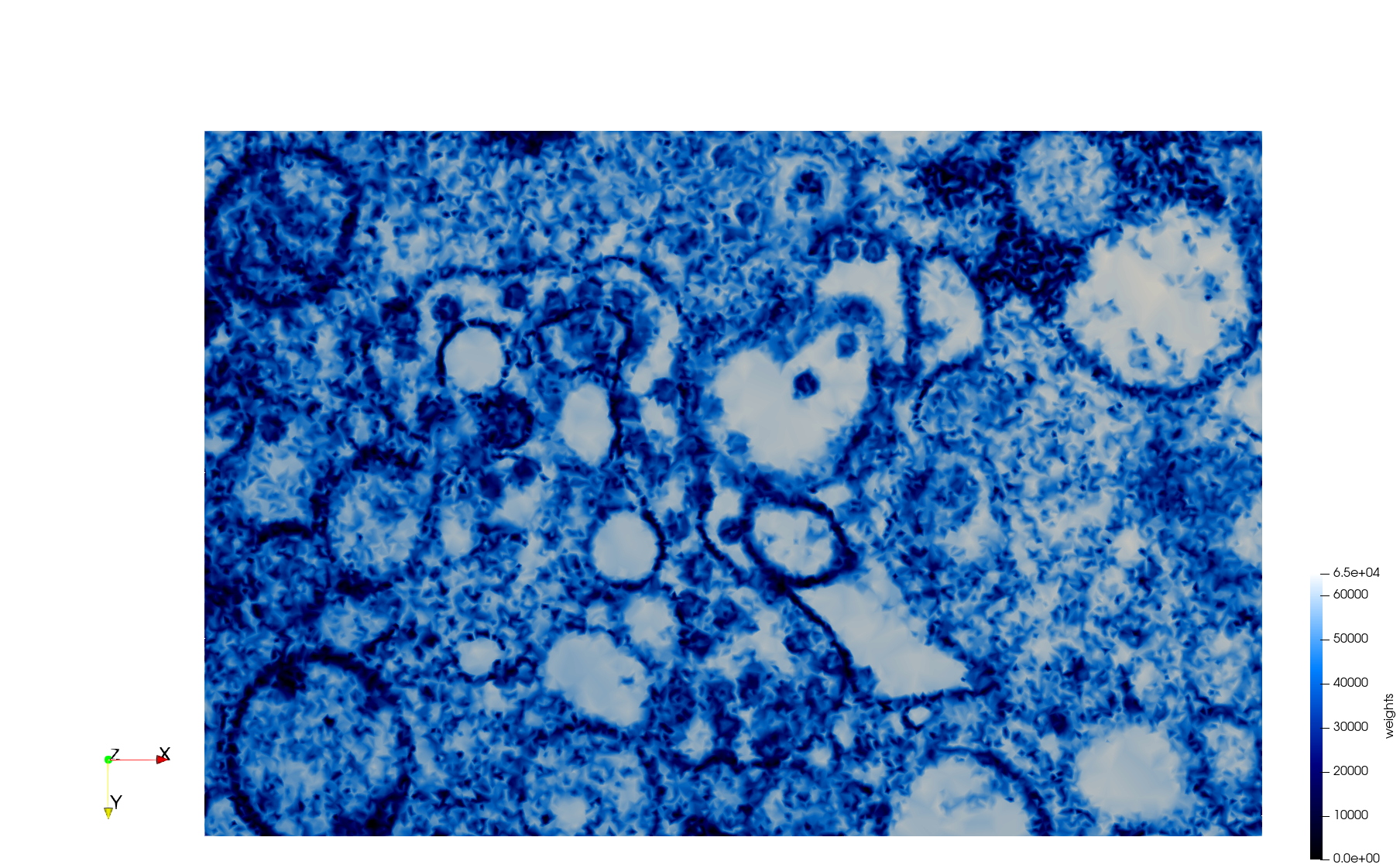
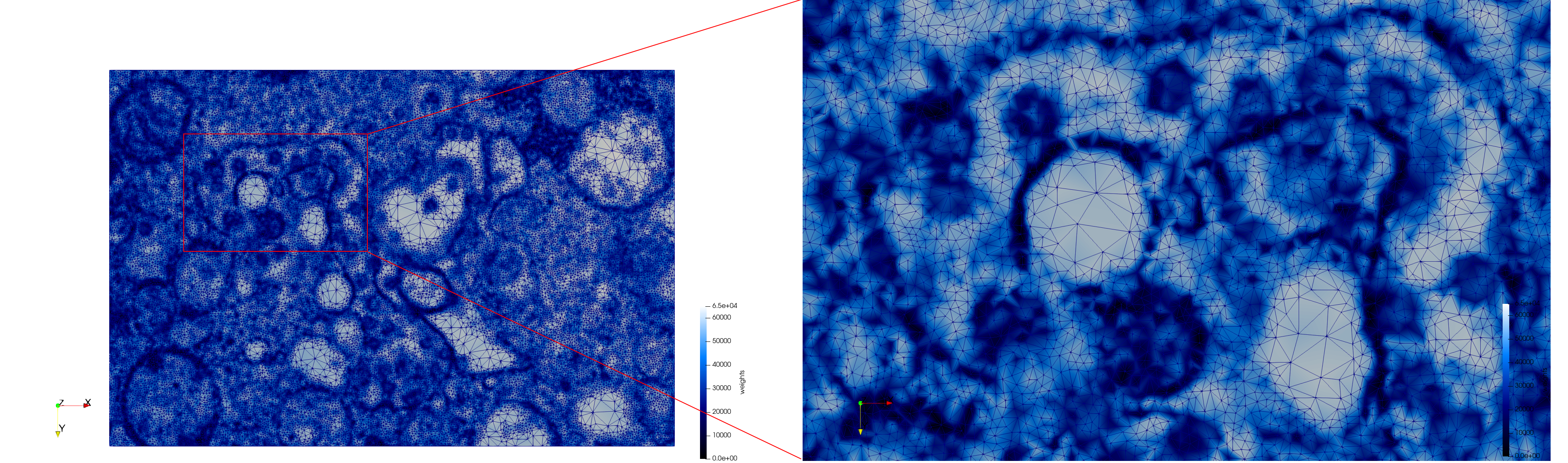
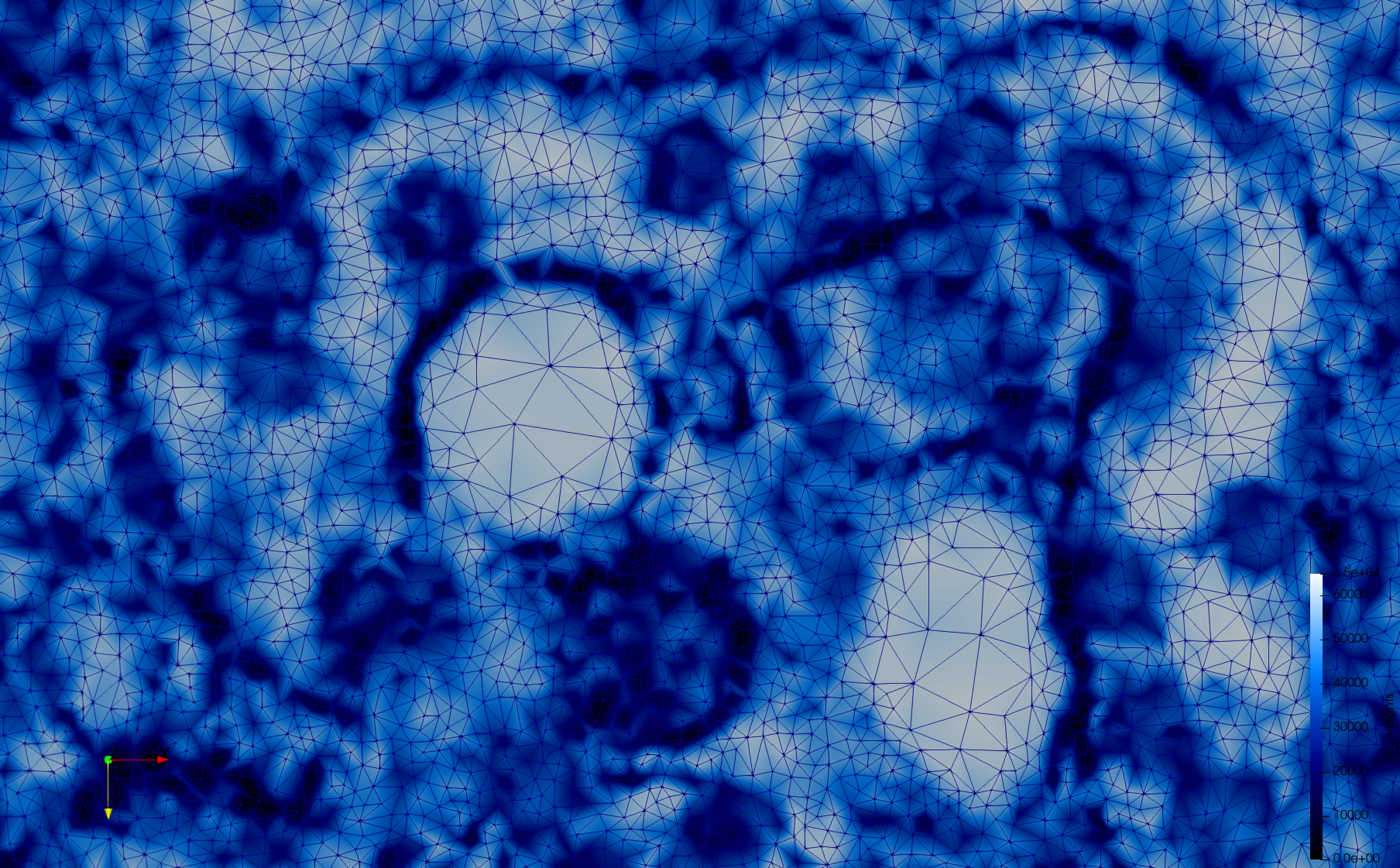
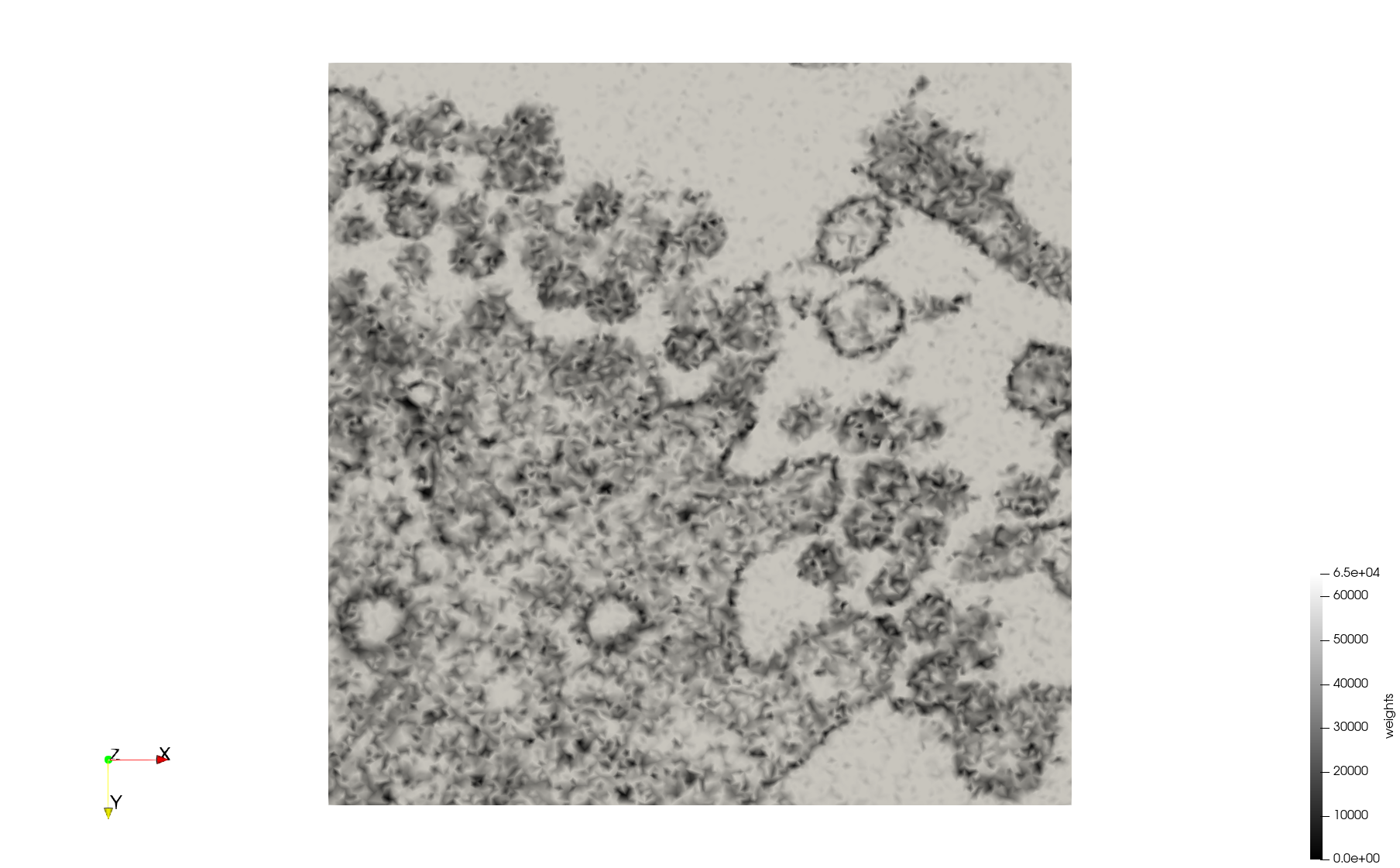
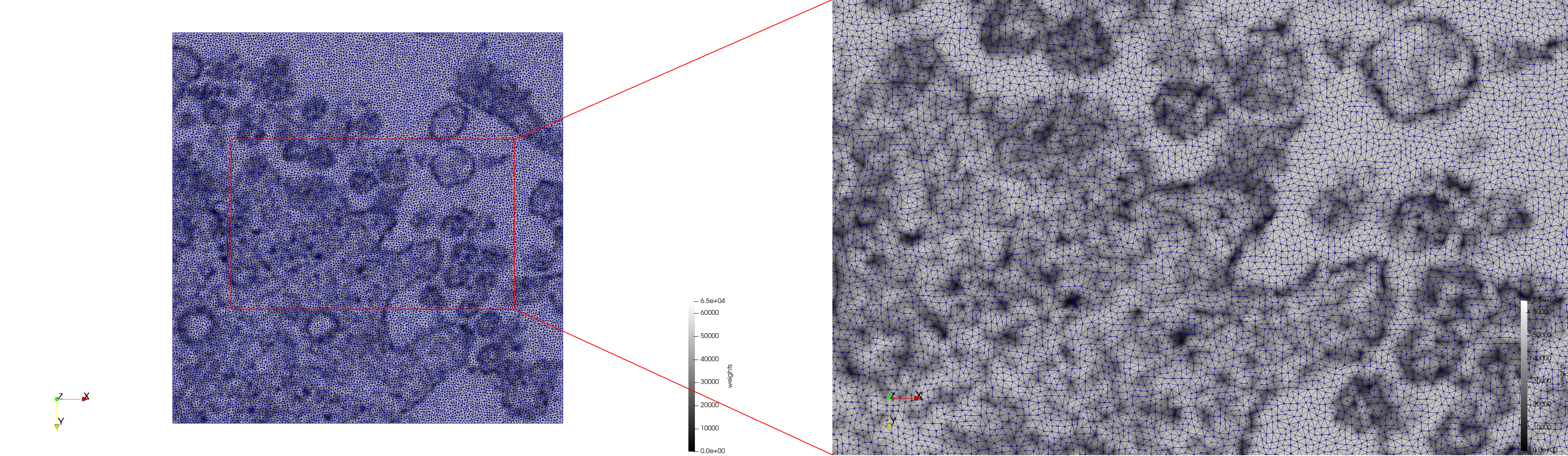
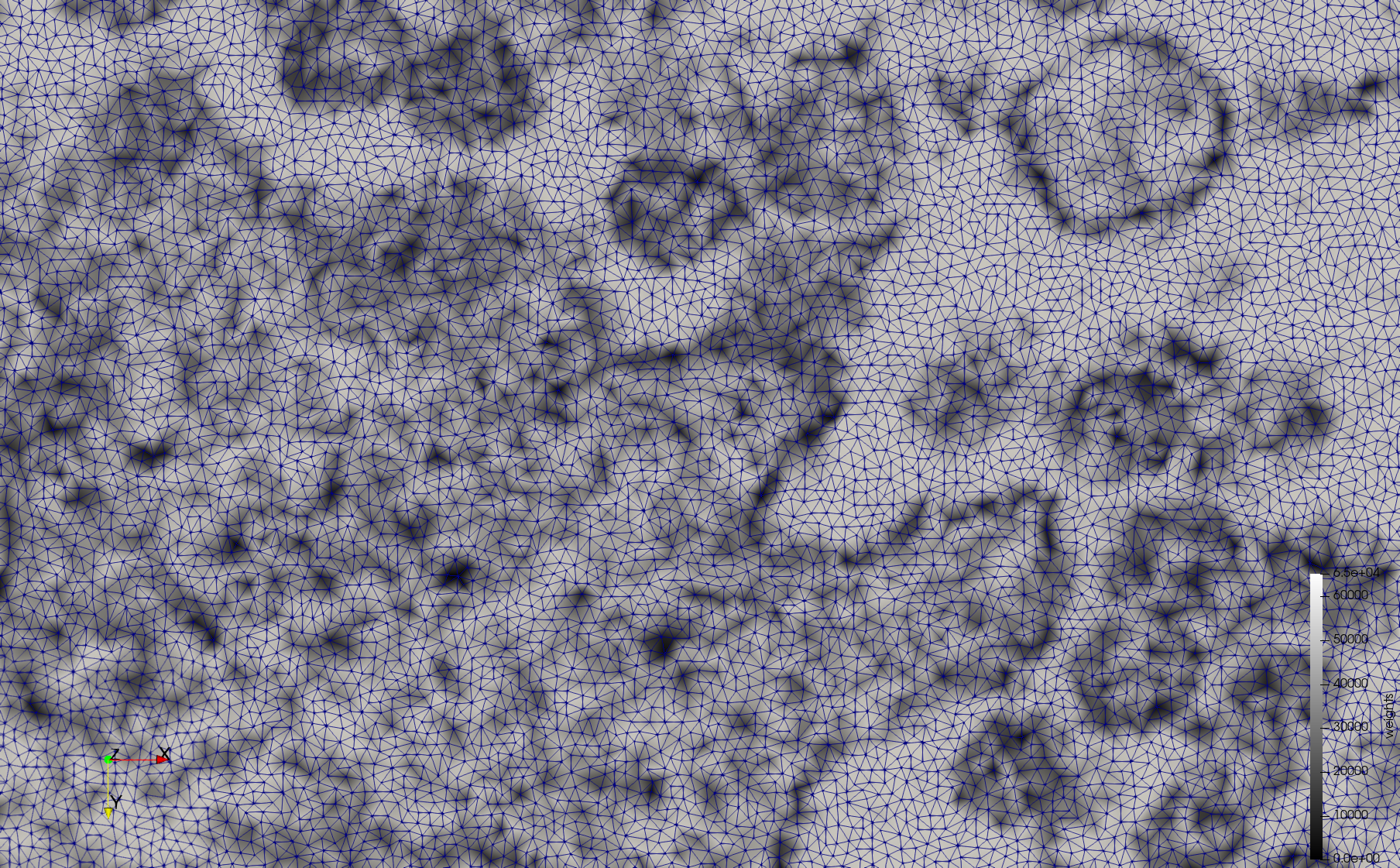
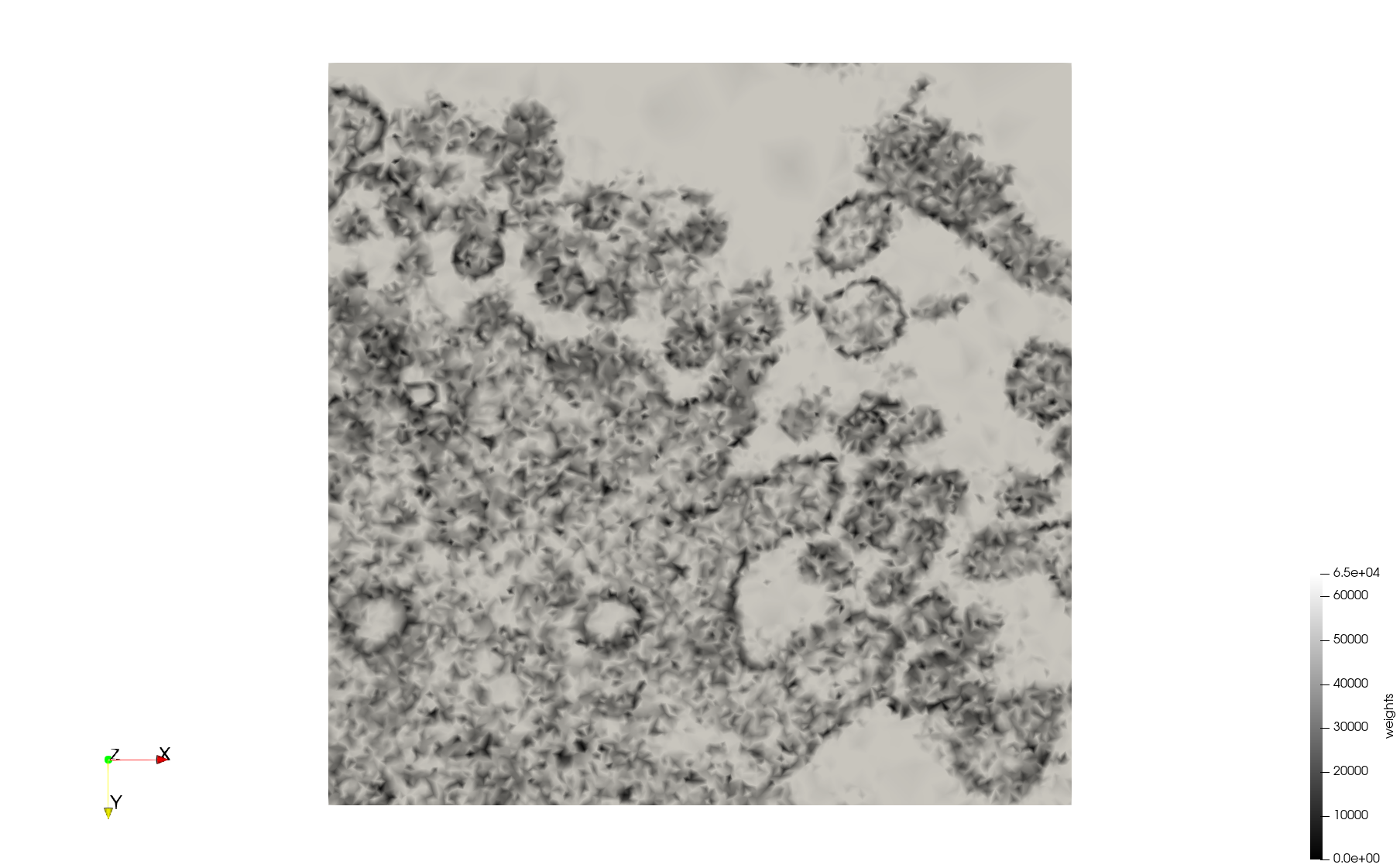
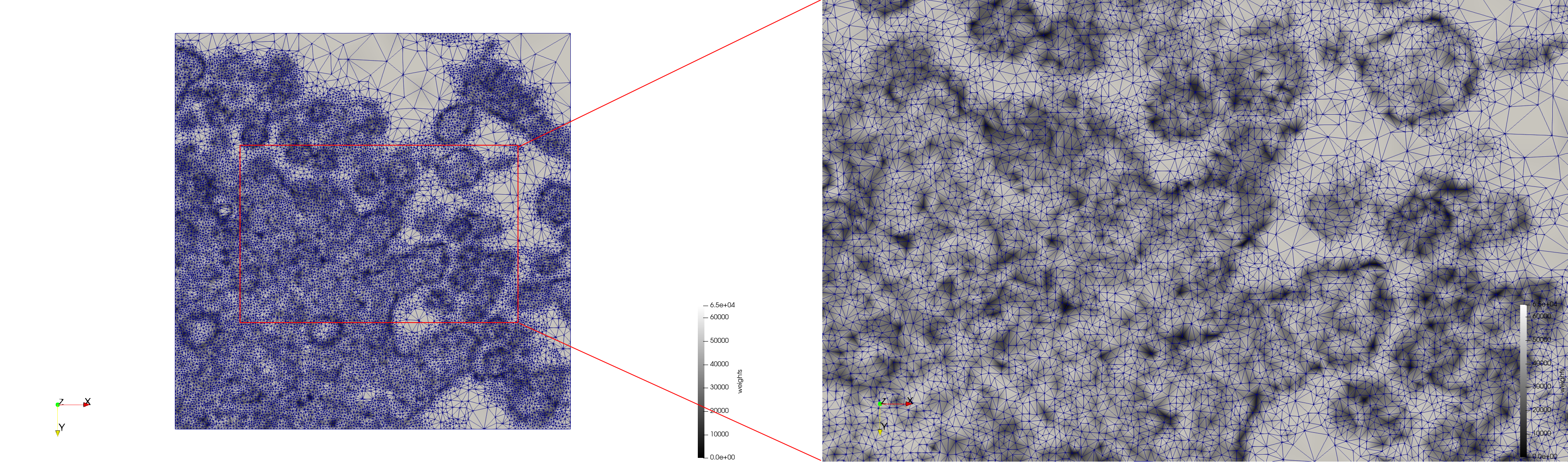
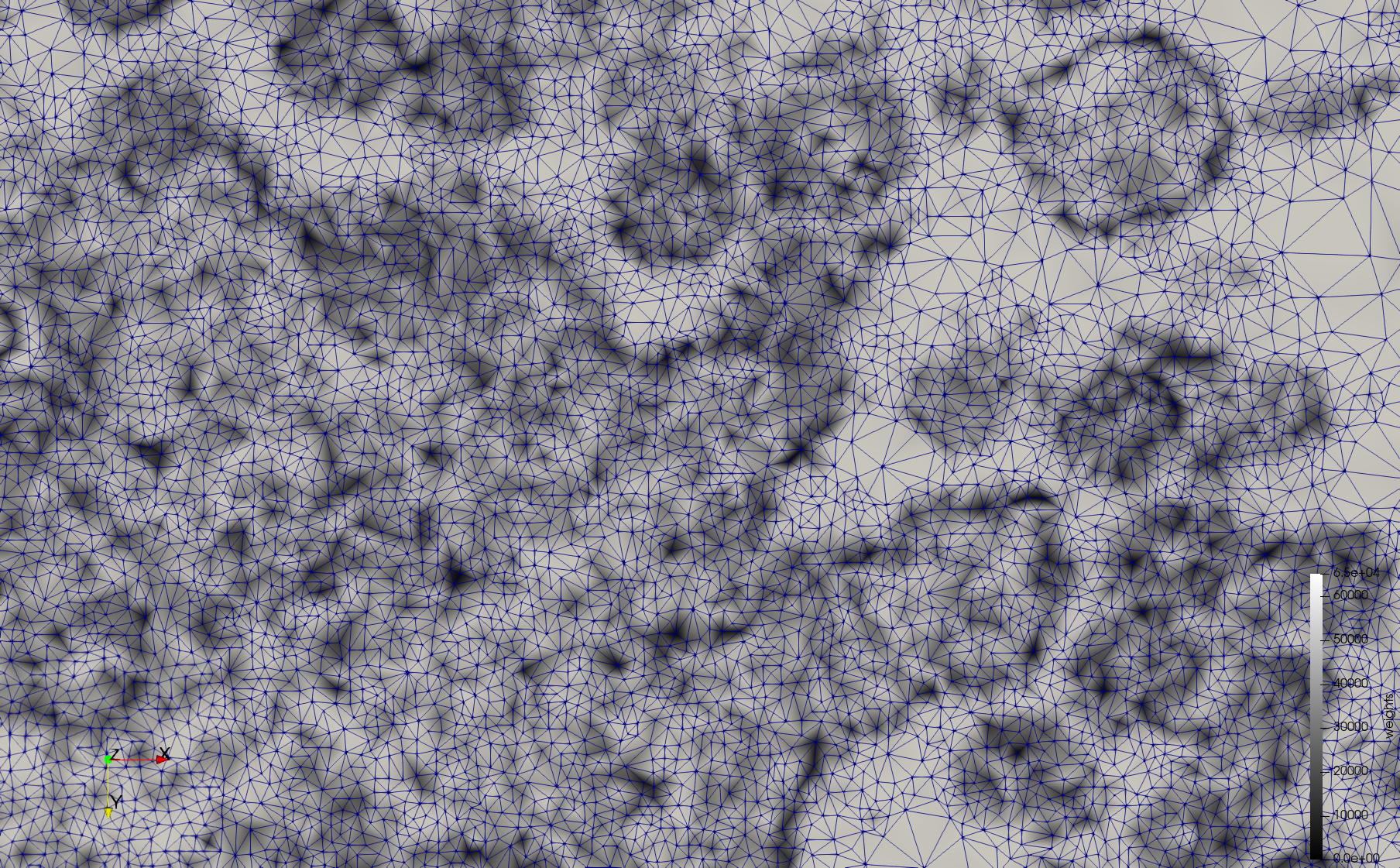
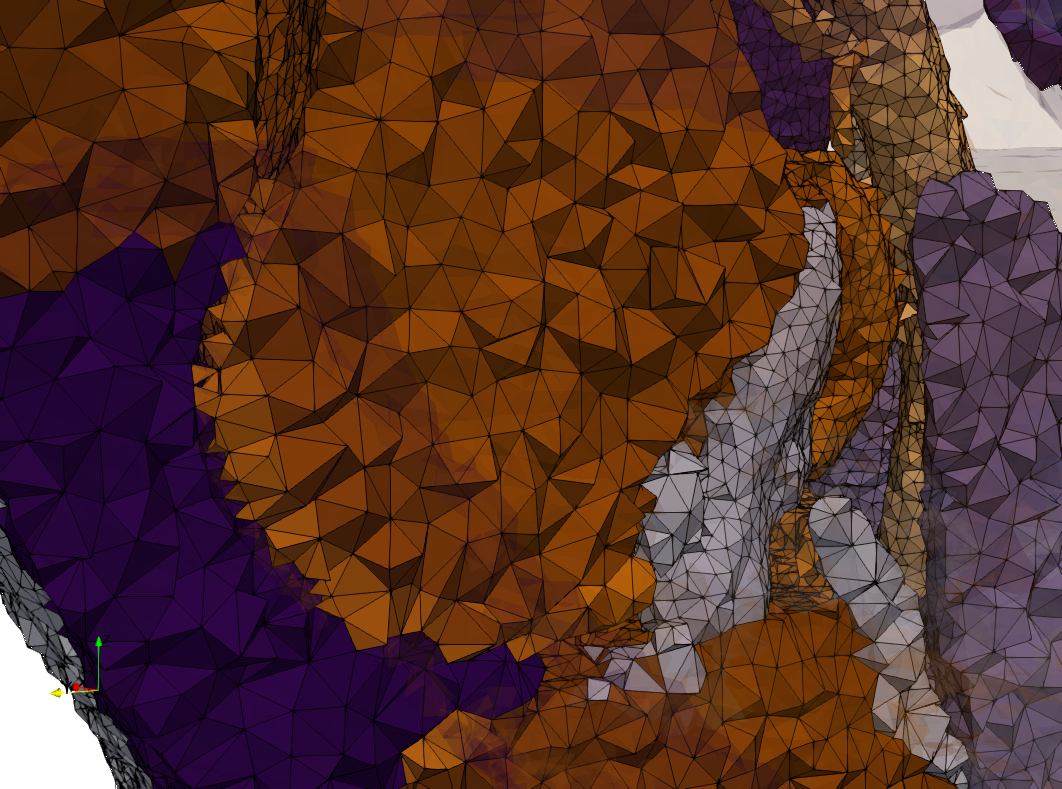
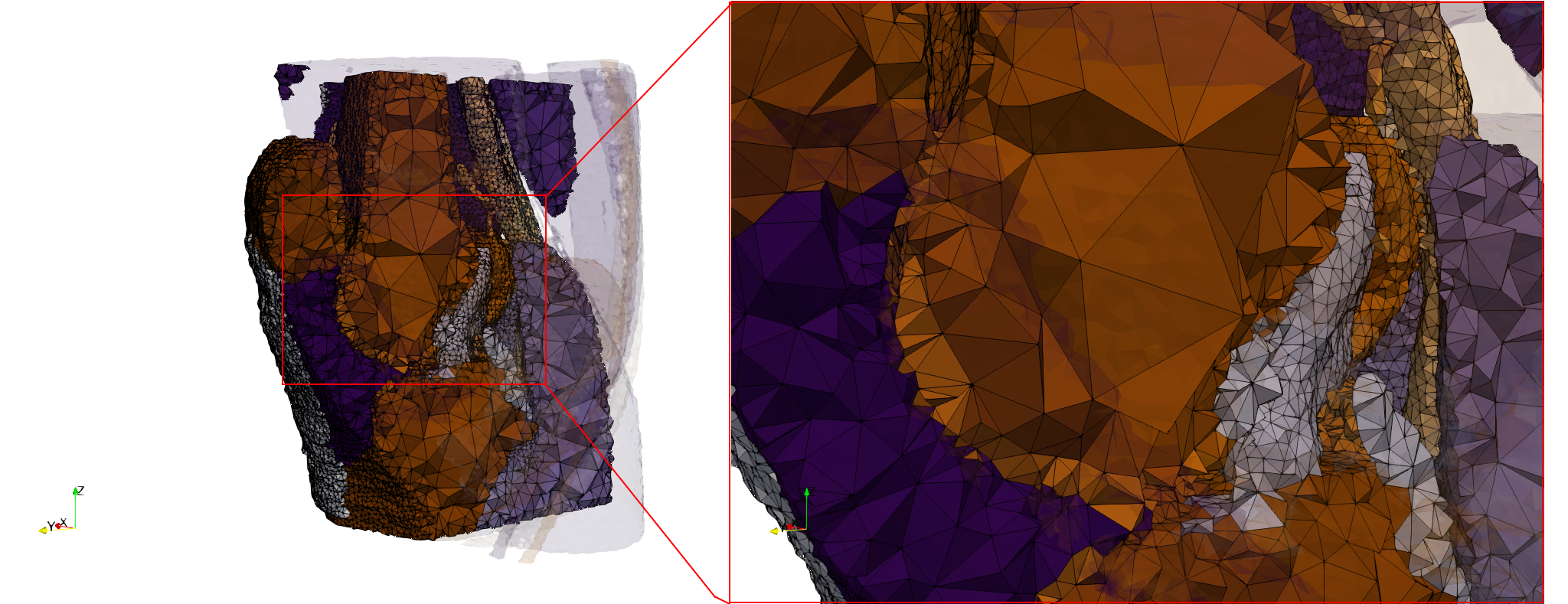
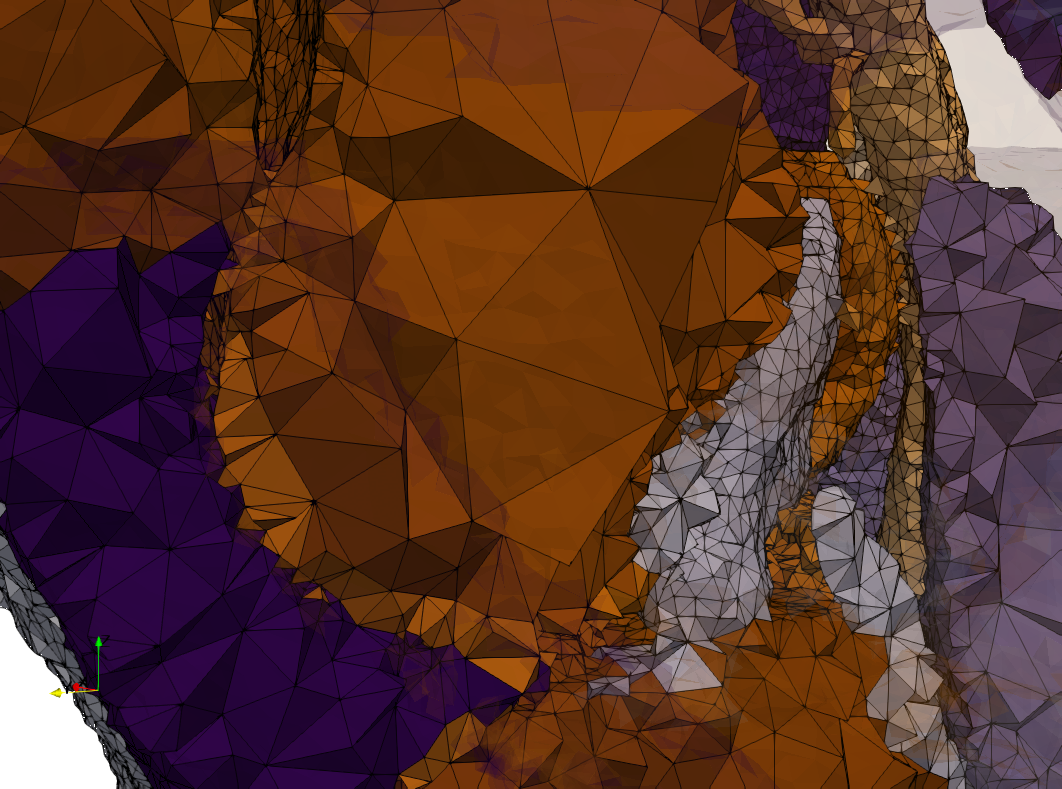
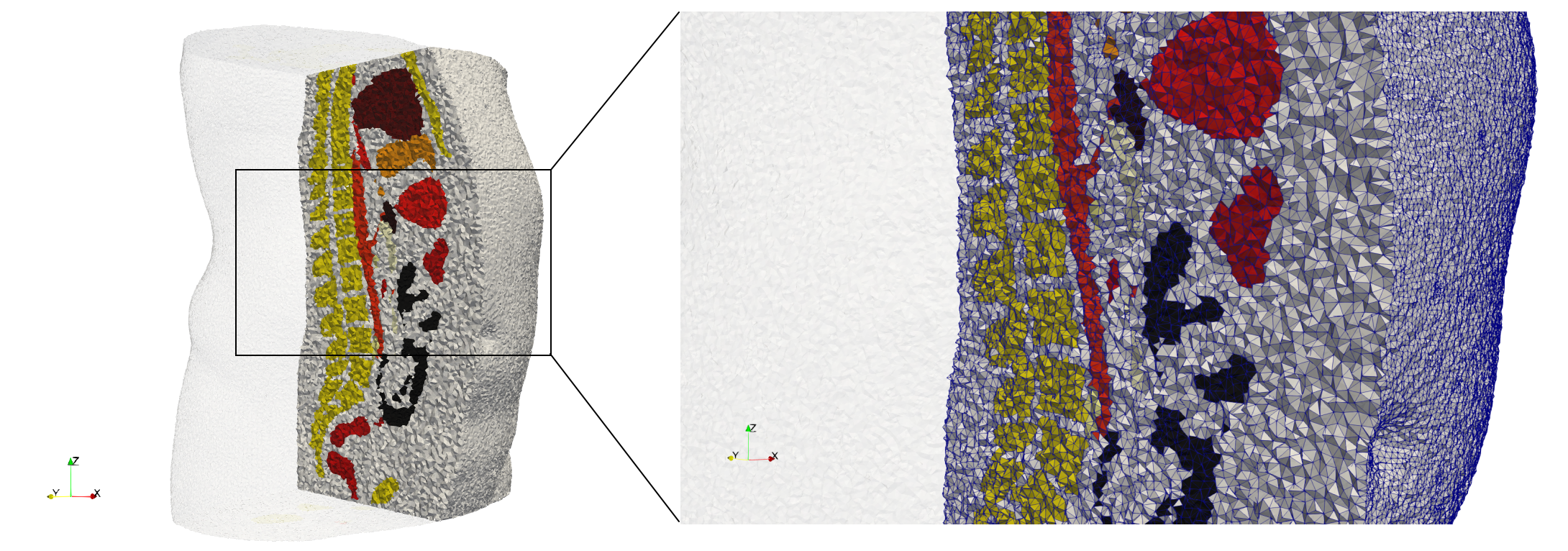
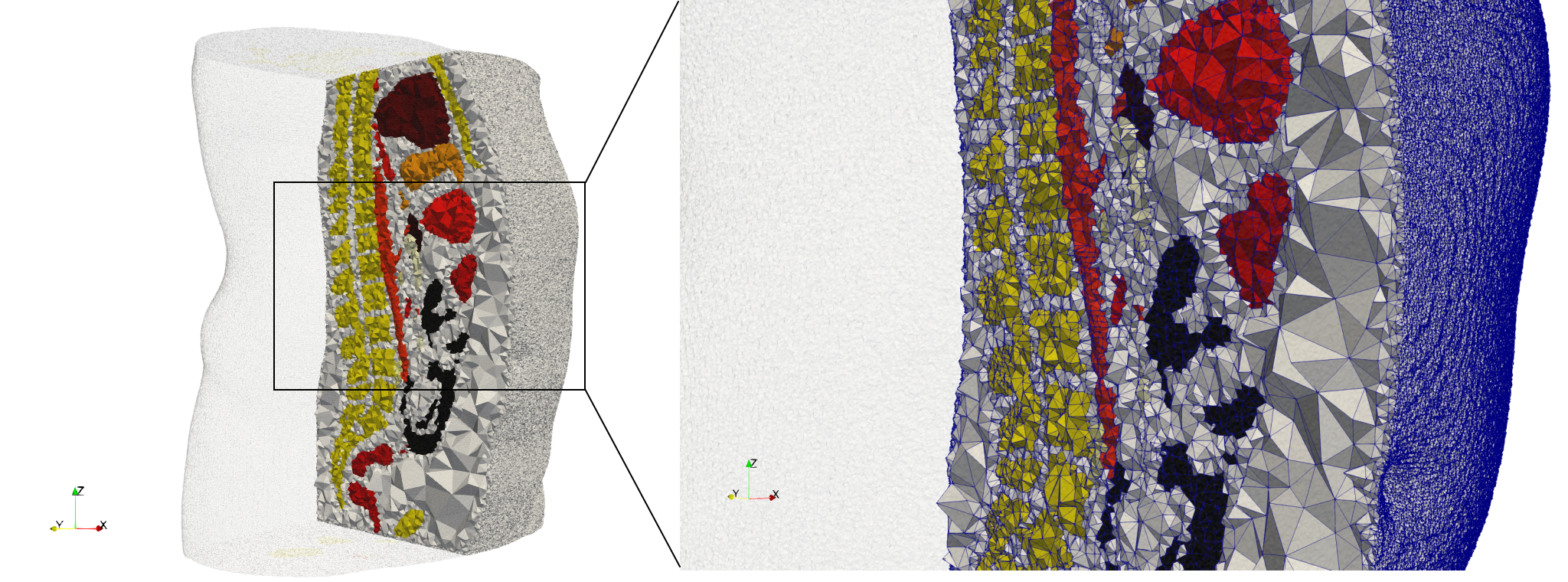
COVID-19-Spike-Glycoprotein-6vsb
- Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042)
- Uniform with Delta = 0.5: 5,731,833 tetrahedra
- Graded with Delta = 0.4: 3,794,223 tetrahedra
[[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4.png] [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip.png] [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip,zoom.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip,zoom.png]
[[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.png] [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip.png] [[File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip,zoom.png|500px|COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip,zoom.png]
Commands to generate meshes :
Uniform with Delta = 0.4: [ Output Mesh]
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4.vtk
Graded with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --volume-grading --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.vtk