Difference between revisions of "Medical Imaging Example Meshes"
Spyridon97 (talk | contribs) (→COVID-19-23354) |
Spyridon97 (talk | contribs) (→2D Example Meshes) |
||
| Line 2: | Line 2: | ||
=2D Example Meshes= | =2D Example Meshes= | ||
The directory containing the 2D input data is located in the 2D folder of [https://odu.box.com/s/wd0i18giti2zo330y19xhkqu8wjzgf7j Medical_Imaging_Data]. | The directory containing the 2D input data is located in the 2D folder of [https://odu.box.com/s/wd0i18giti2zo330y19xhkqu8wjzgf7j Medical_Imaging_Data]. | ||
| − | |||
| − | |||
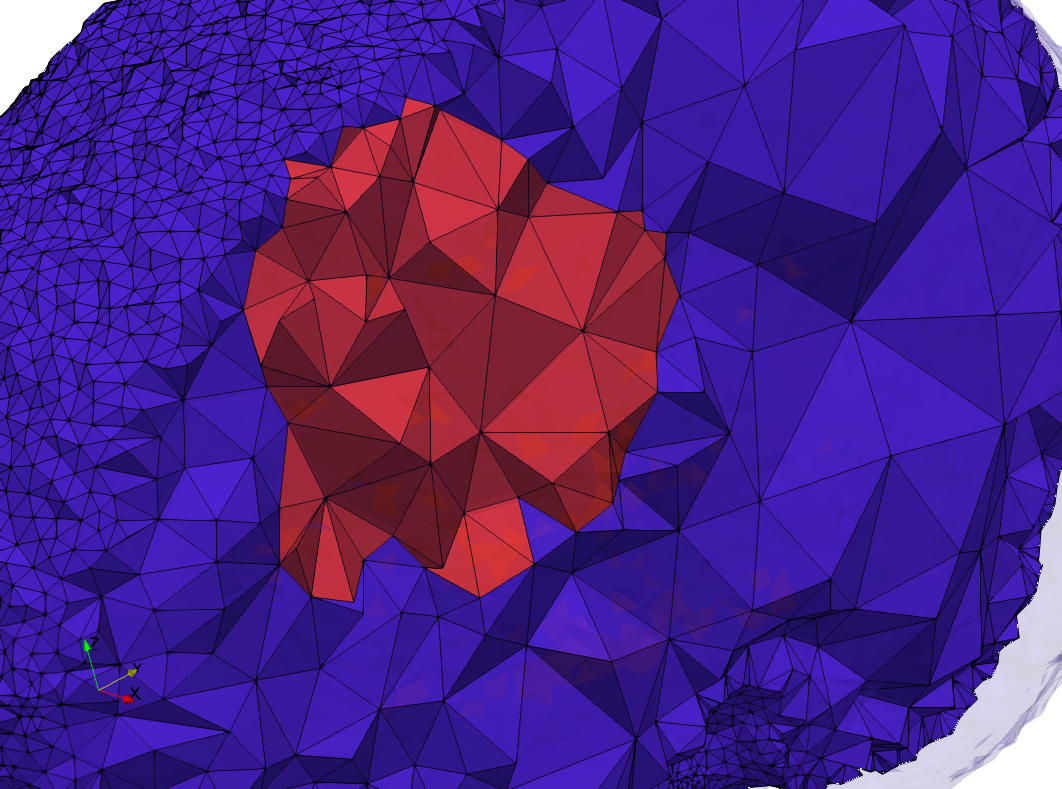
==COVID-19-23354== | ==COVID-19-23354== | ||
Revision as of 19:46, 14 March 2020
Contents
2D Example Meshes
The directory containing the 2D input data is located in the 2D folder of Medical_Imaging_Data.
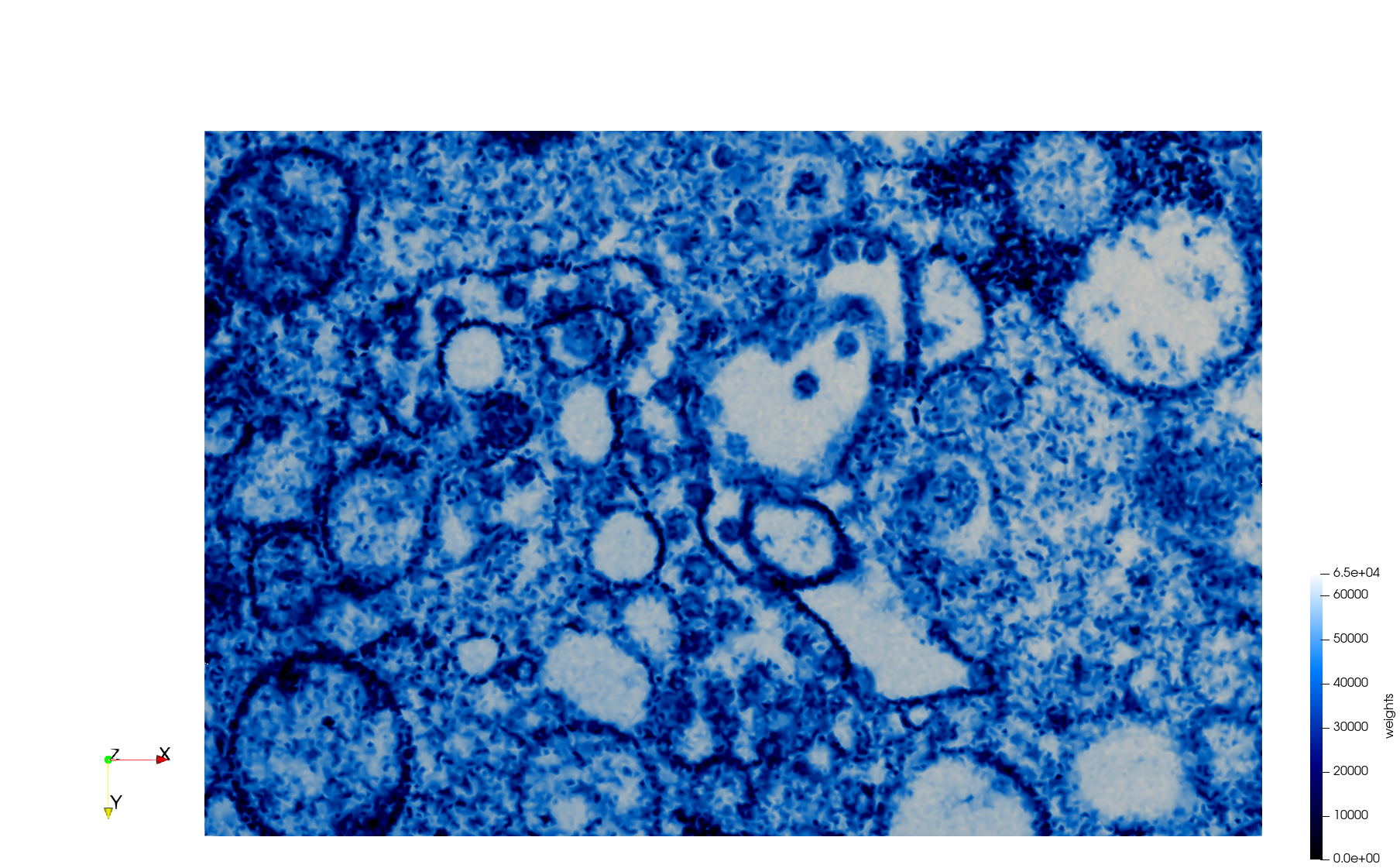
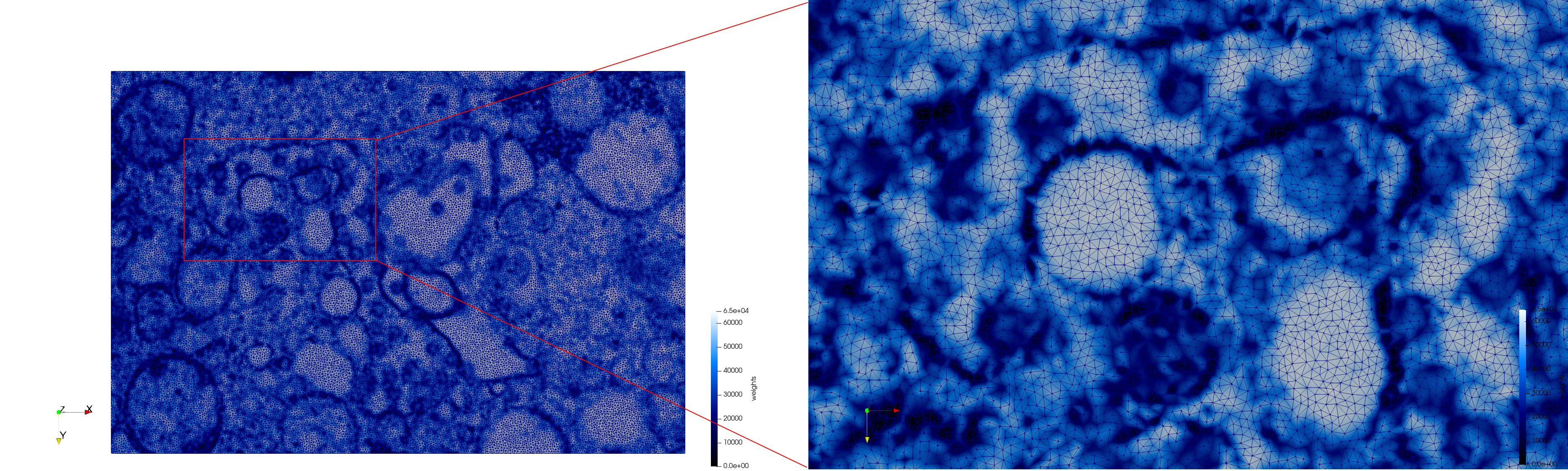
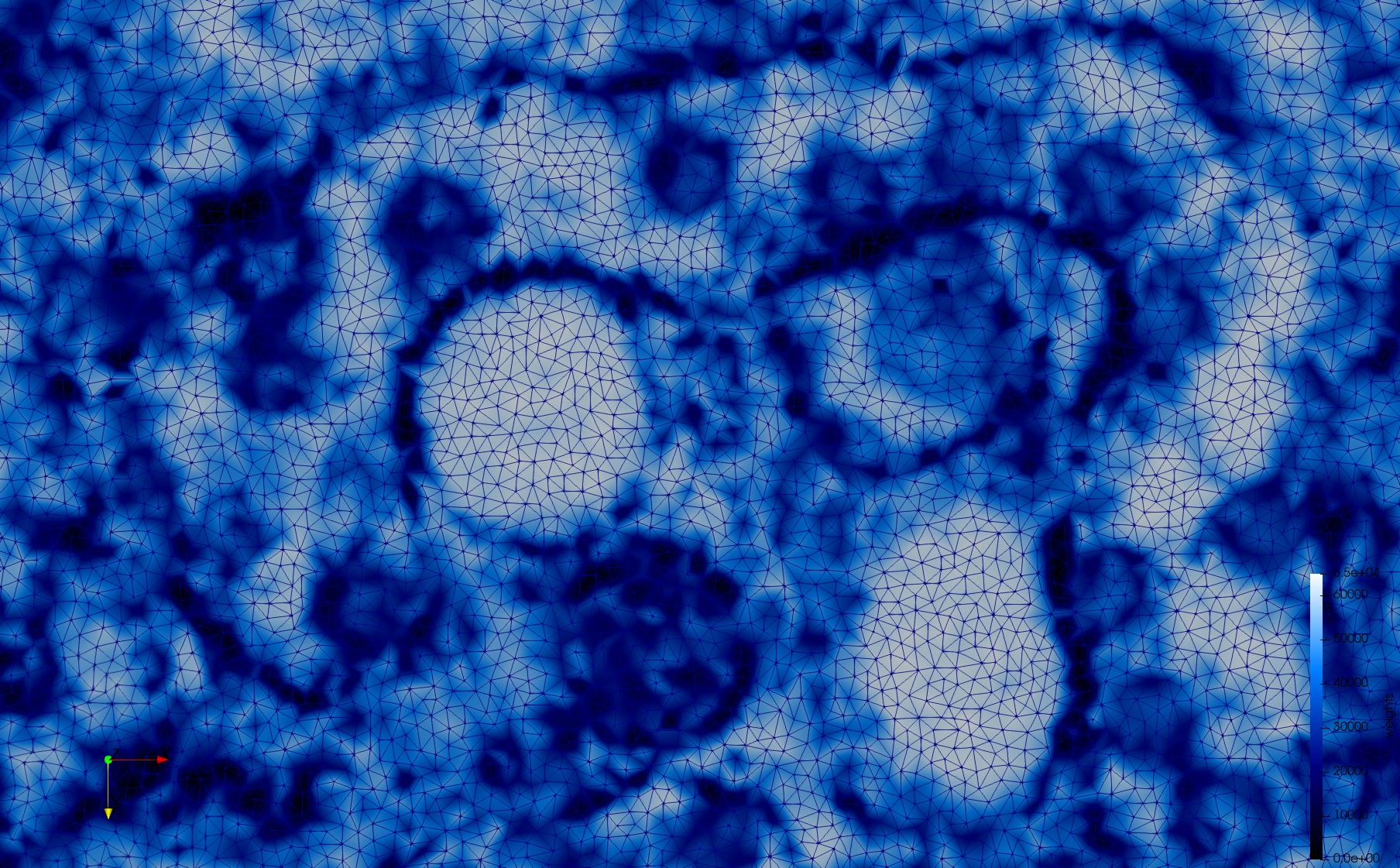
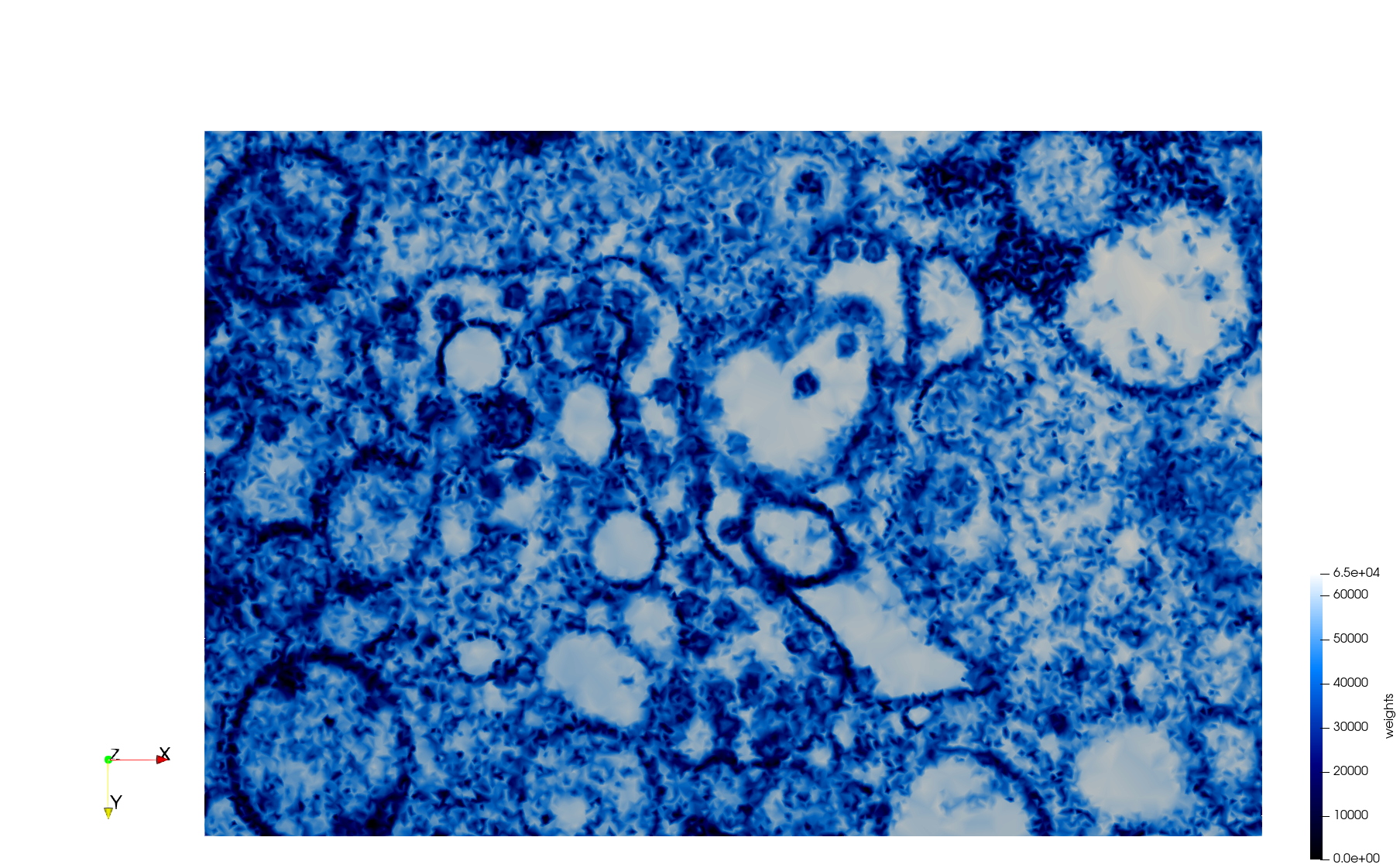
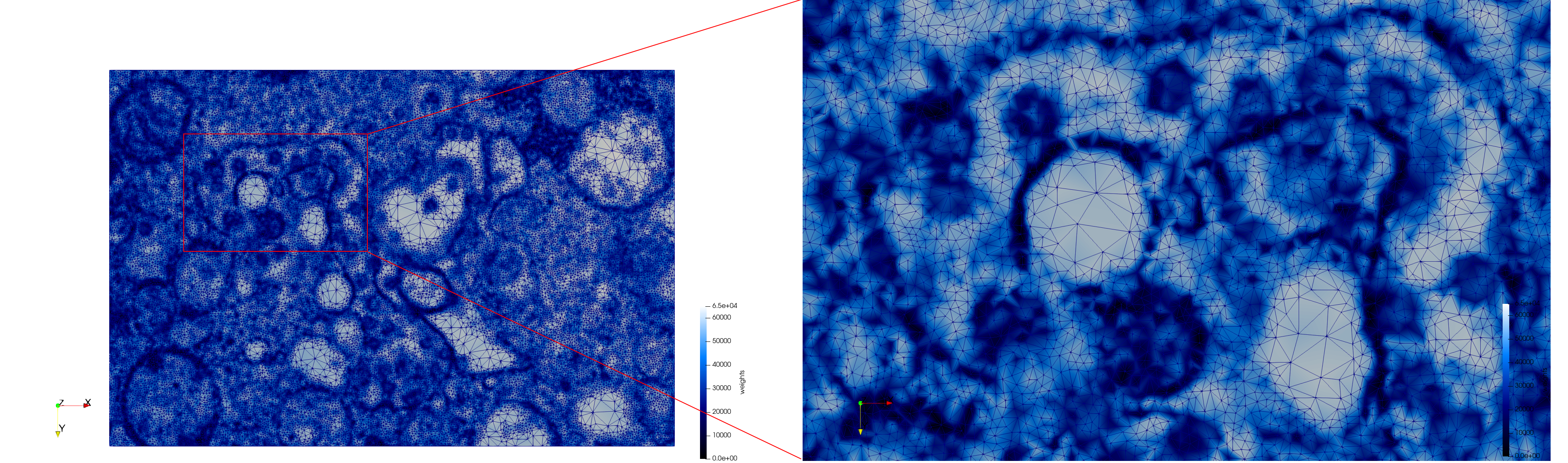
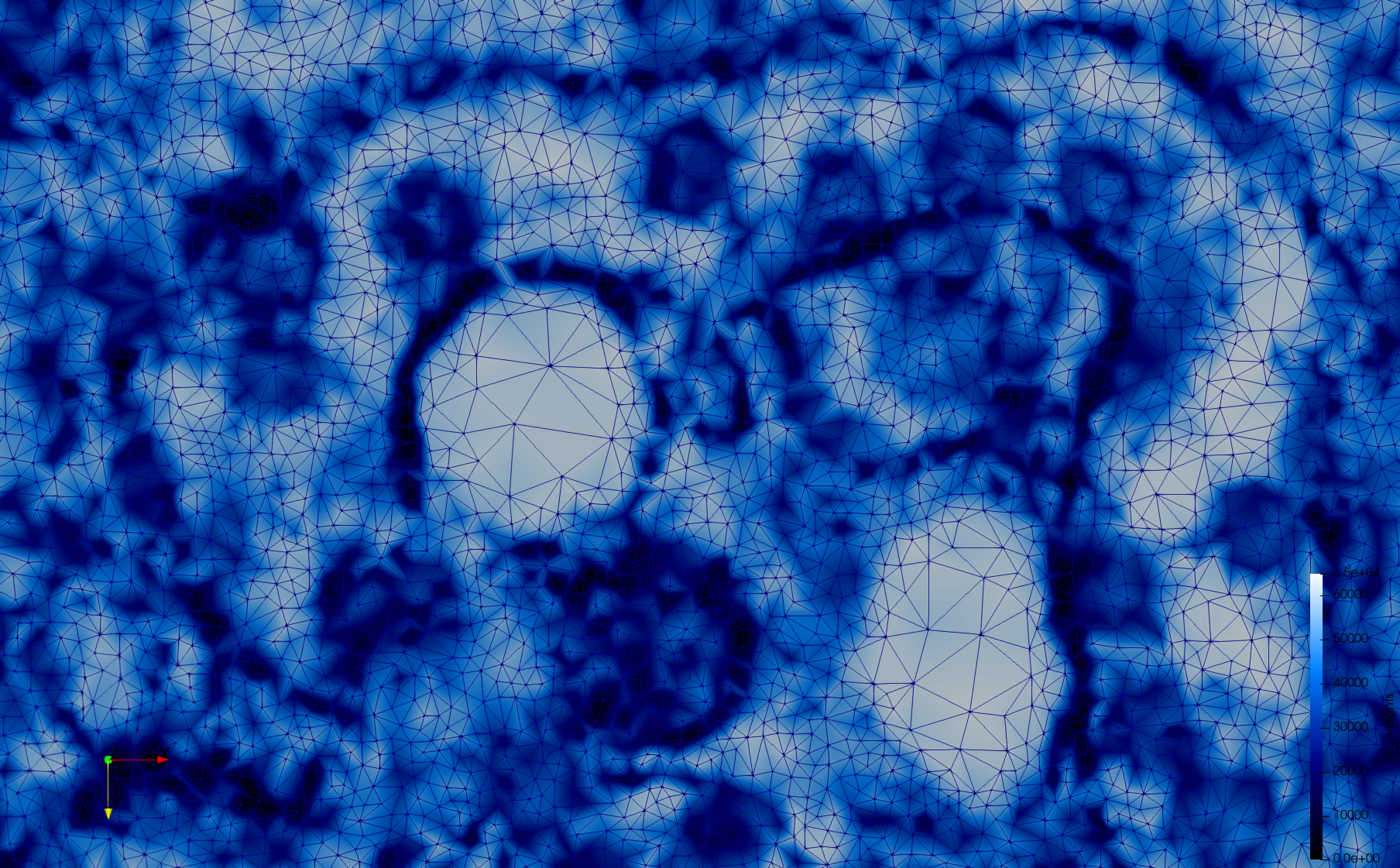
COVID-19-23354
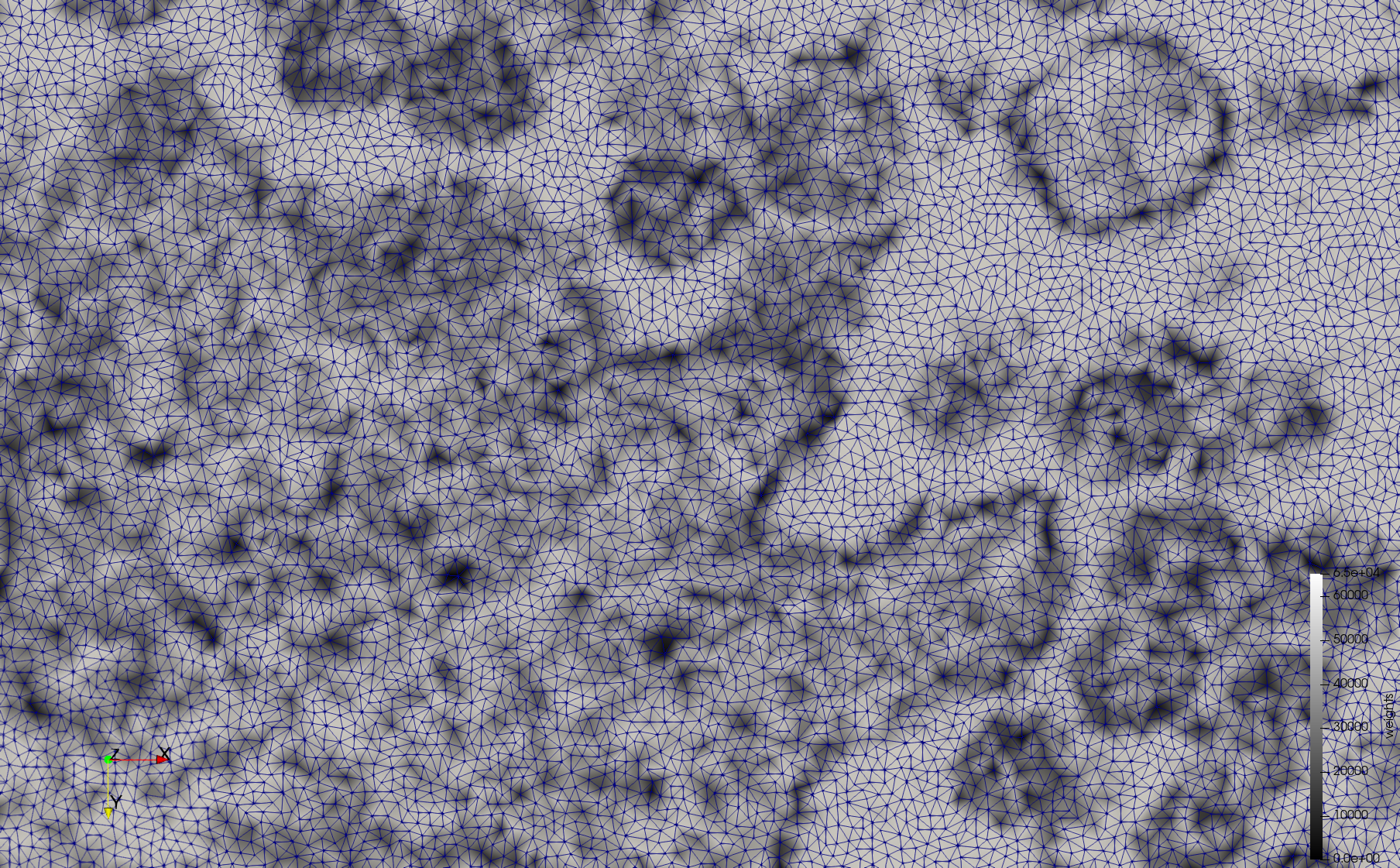
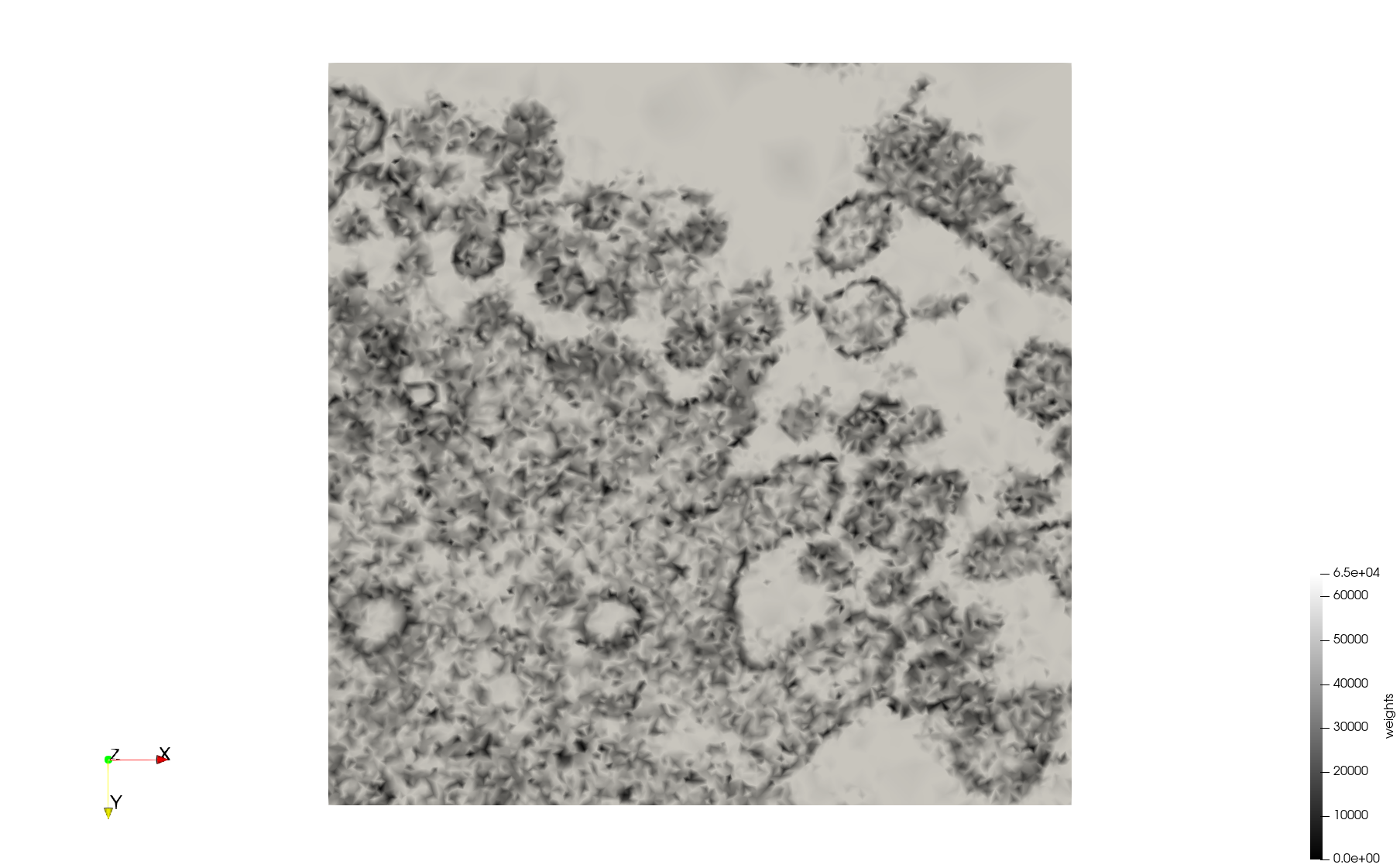
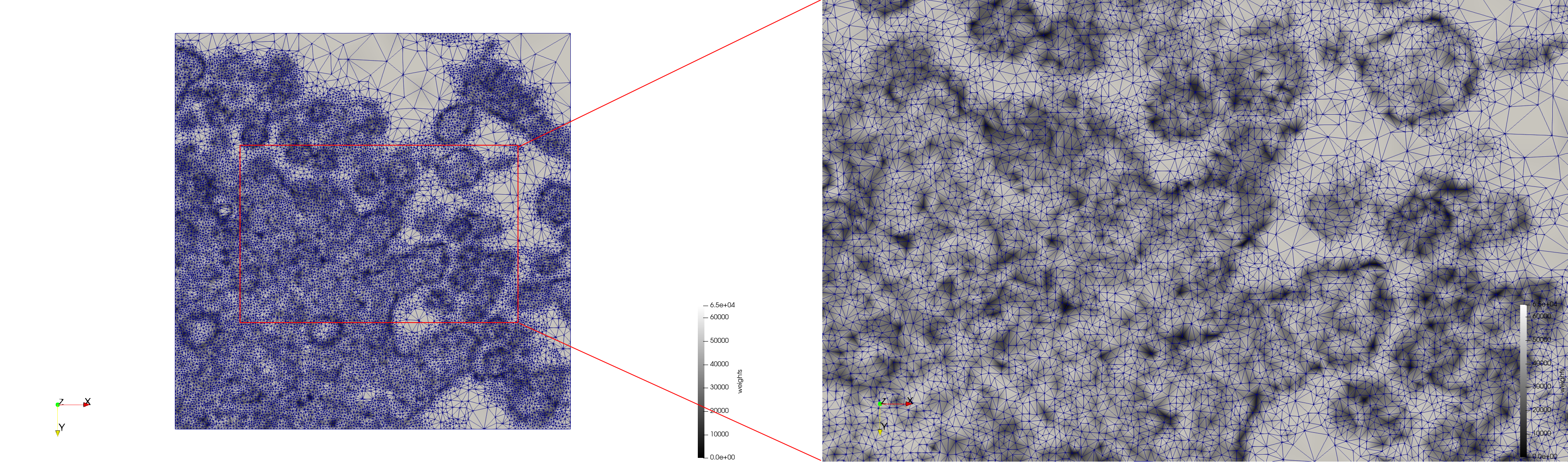
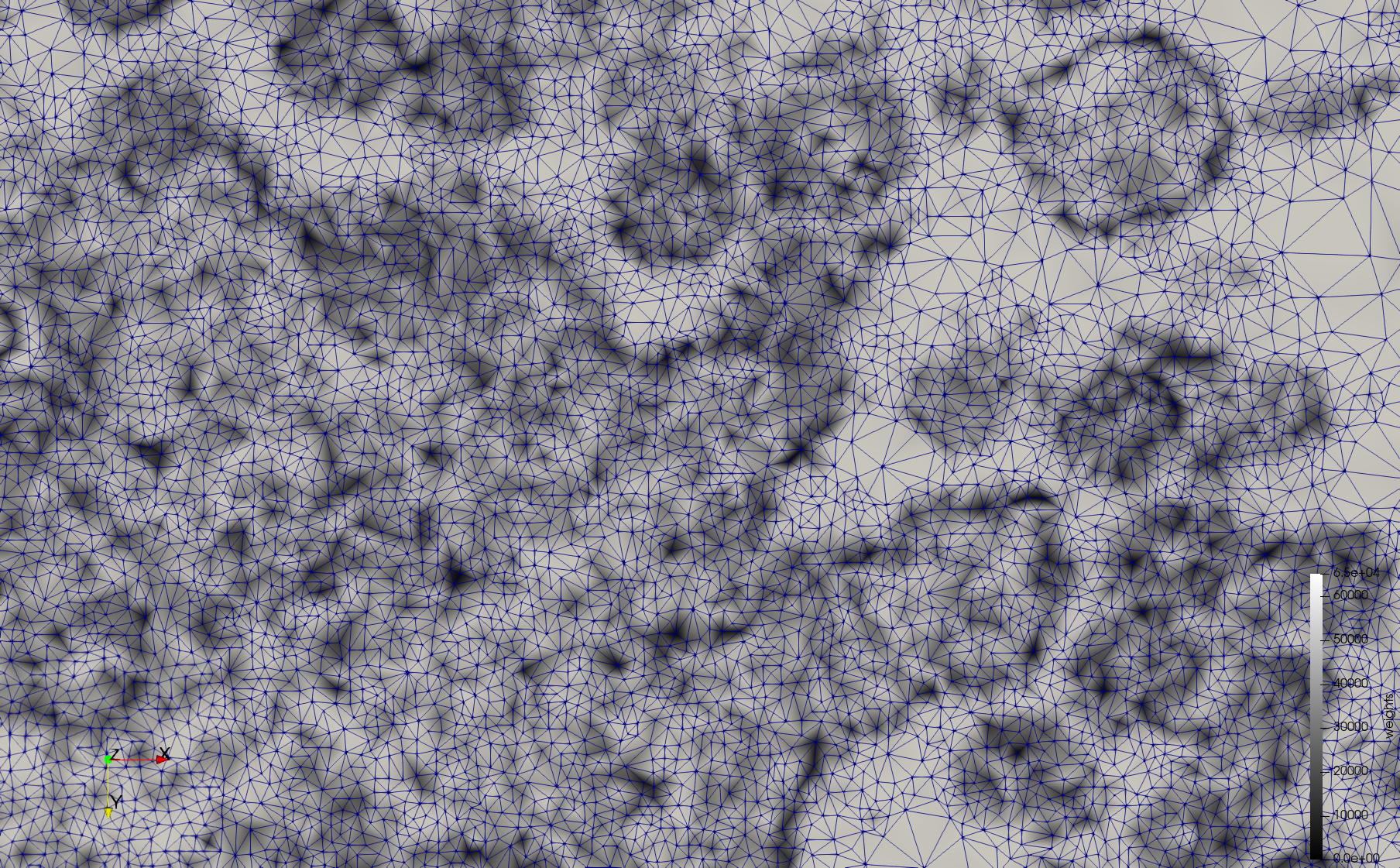
- Input image : Dimensions (3,000x2,000) with spacing (1x1)
- Uniform with Min-Edge = 15: 82,981 triangles
- Adaptive with Min-Edge = 15: 67,920 triangles
Commands to generate meshes :
Uniform with Min-Edge = 15 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --uniform --min-edge 15 --output ./COVID-19-23354,uniform,e=15.vtk
Adaptive with Min-Edge = 15 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --min-edge 15 --output ./COVID-19-23354,w=0.1,e=15.vtk
Mesh generated based on the images published at Centers for Disease Control and Prevention. For the uniform meshes, the edge-size corresponds to 15 pixels. The adaptive meshes were created by controlling the size of the elements based on the difference in the intensity of the pixels.
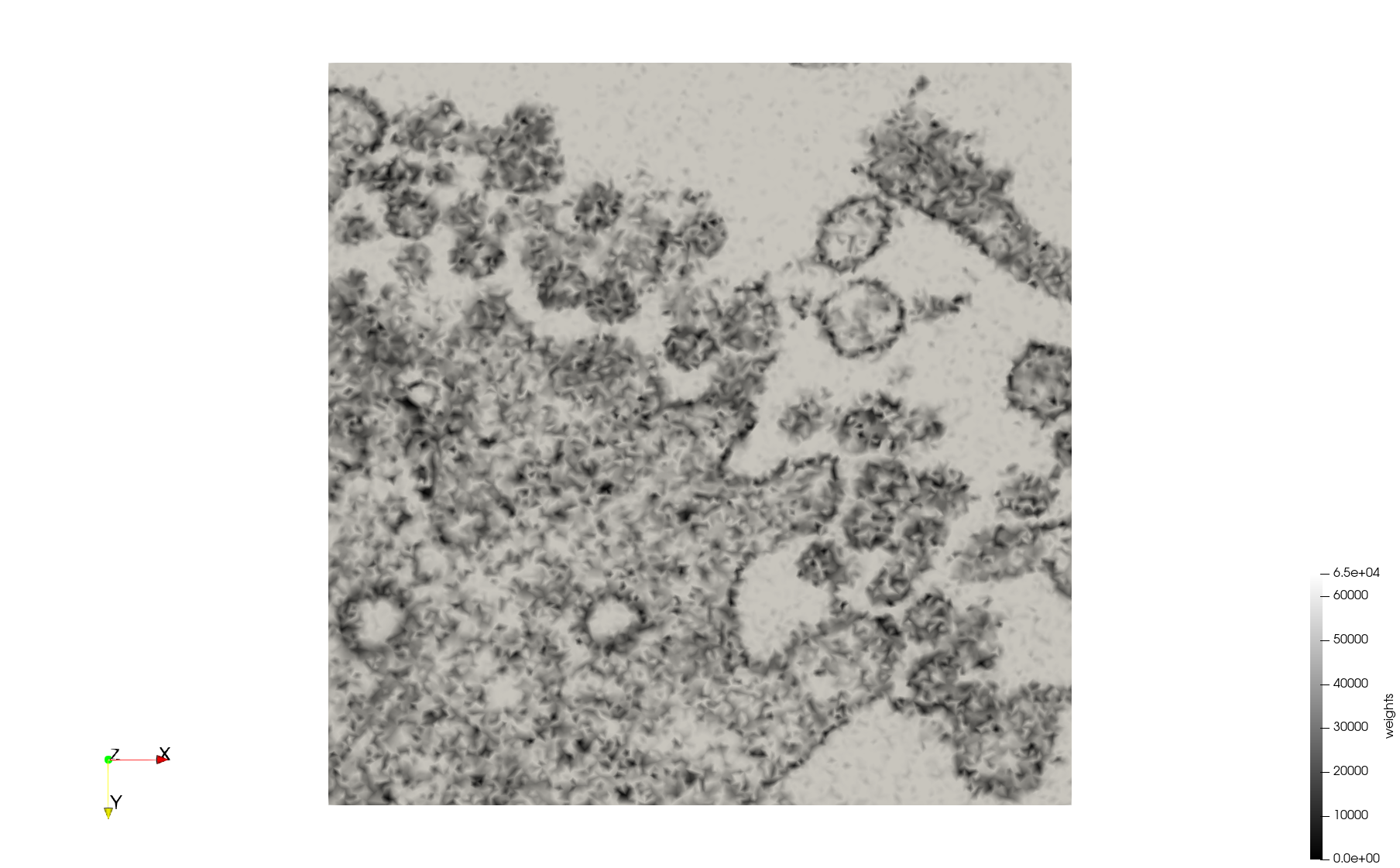
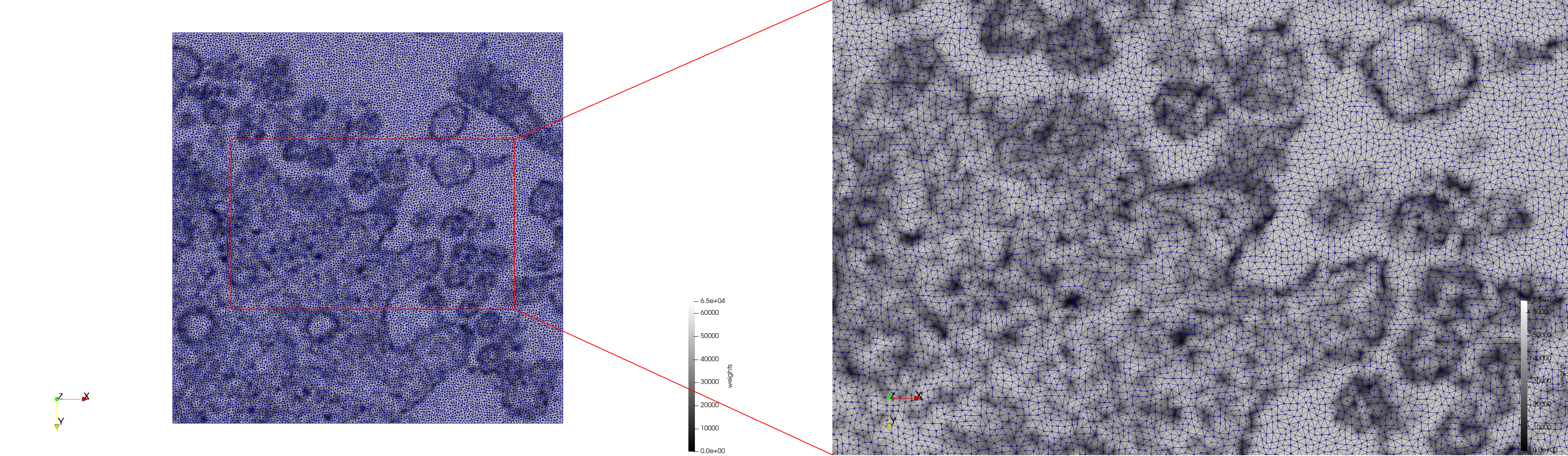
COVID-19-23311
- Input image : Dimensions (2,460x2,460) with spacing (1x1)
- Uniform with Min-Edge = 20: 47,028 triangles
- Adaptive with Min-Edge = 20: 34,080 tetrahedra
Commands to generate meshes :
Uniform with Min-Edge = 20 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --uniform --min-edge 20 --output ./COVID-19-23311,uniform,e=20.vtk
Adaptive with Min-Edge = 20 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --min-edge 20 --output ./COVID-19-23311,w=0.1,e=20.vtk
Mesh generated based on the images published at Centers for Disease Control and Prevention. For the uniform meshes, the edge-size corresponds to 20 pixels. The adaptive meshes were created by controlling the size of the elements based on the difference in the intensity of the pixels.
3D Example Meshes
The directory containing the 3D input data is located in the 3D folder of Medical_Imaging_Data.
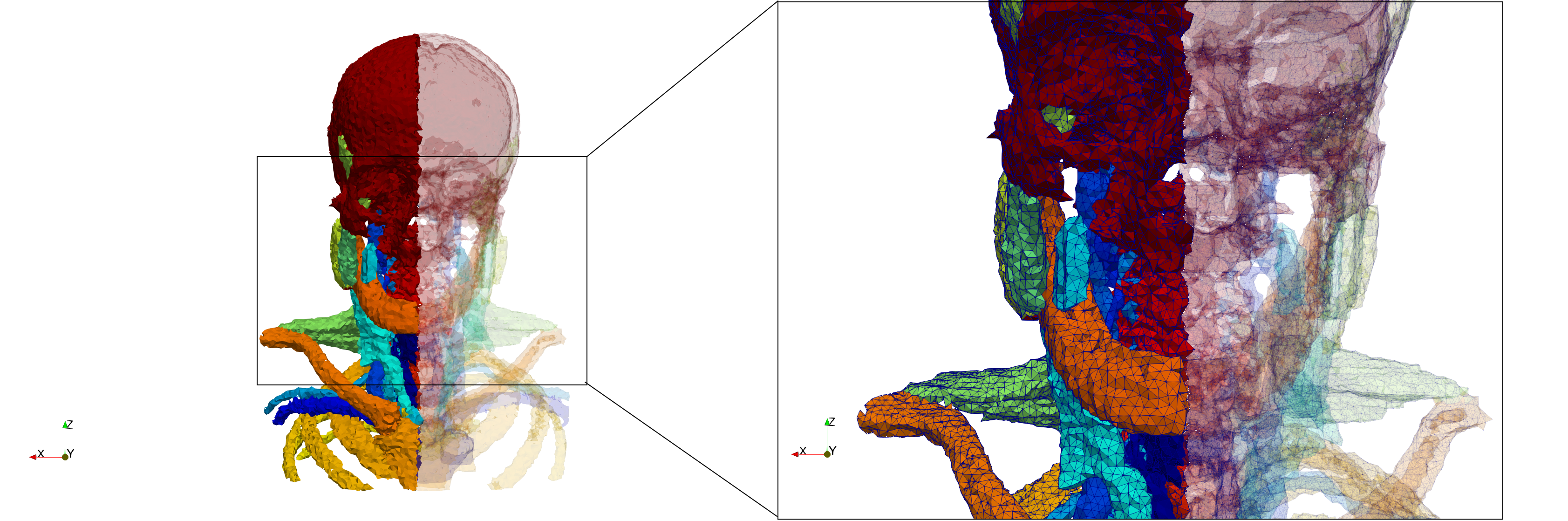
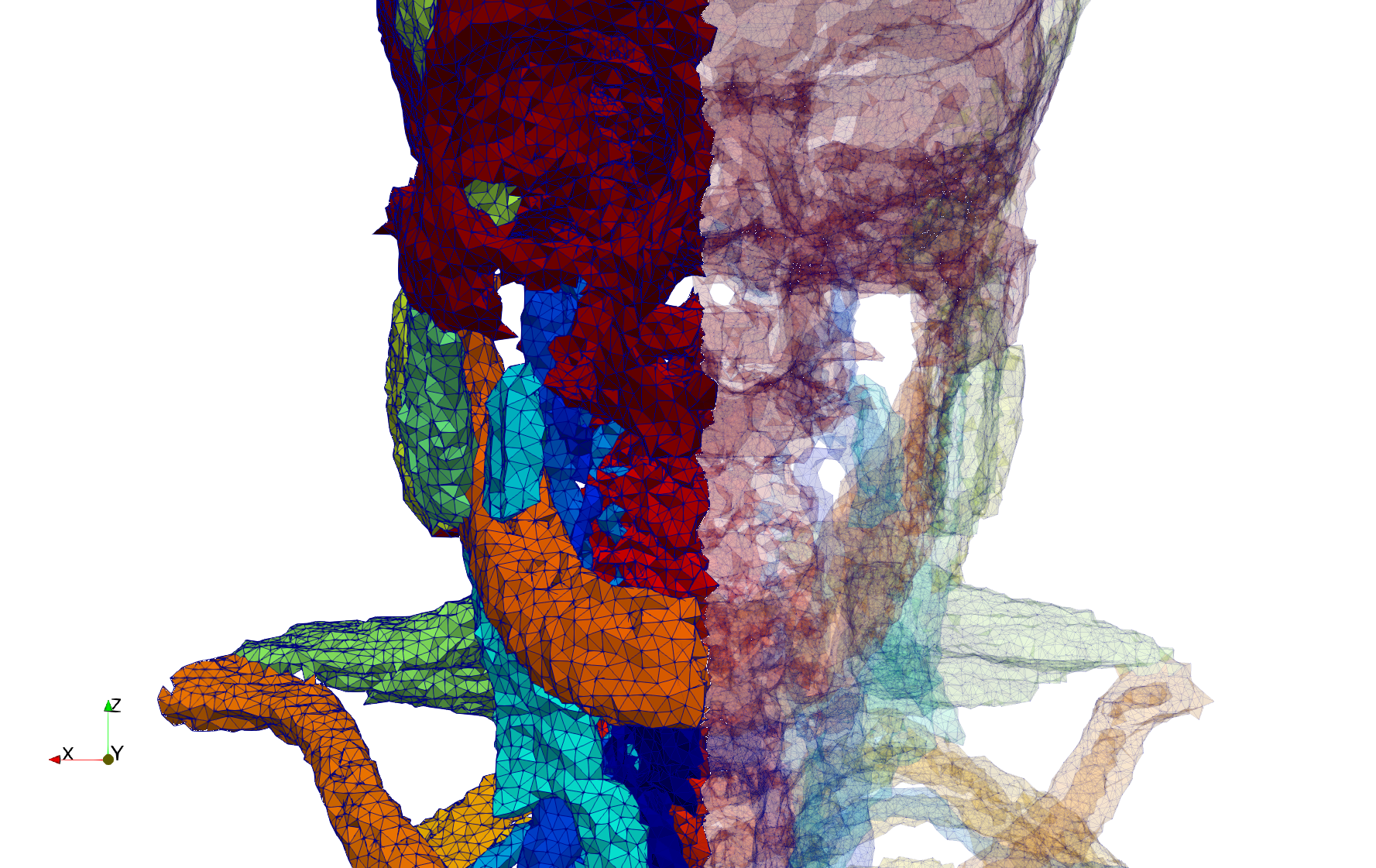
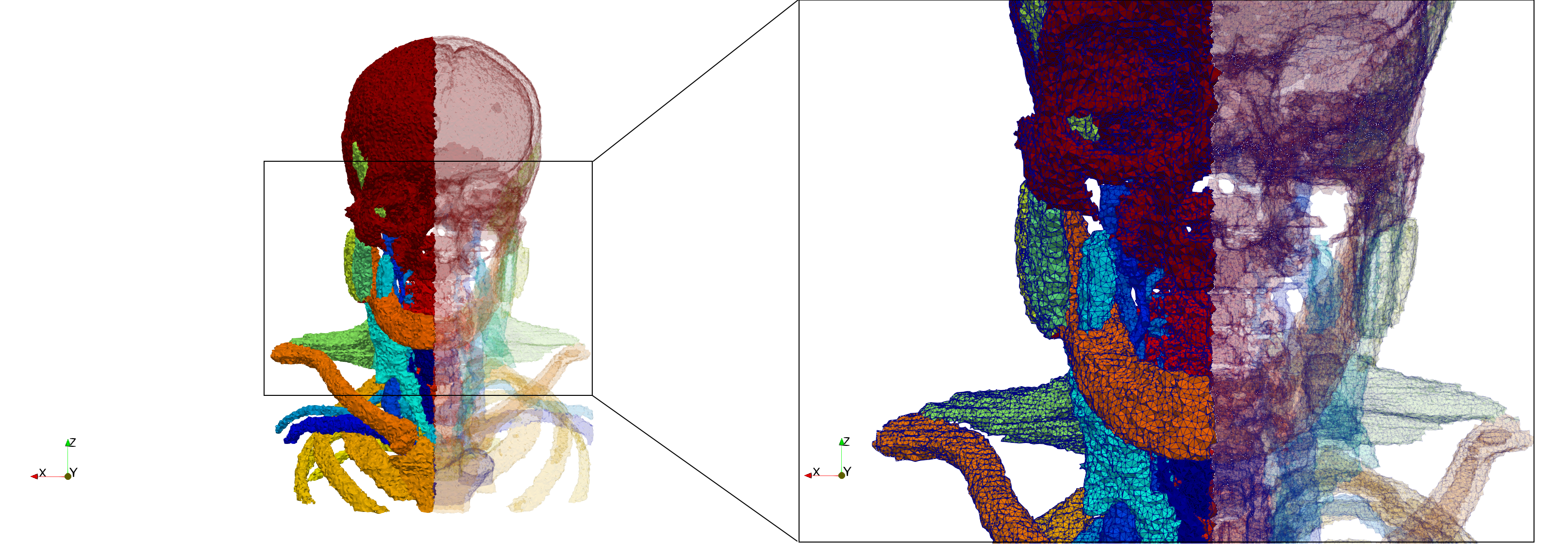
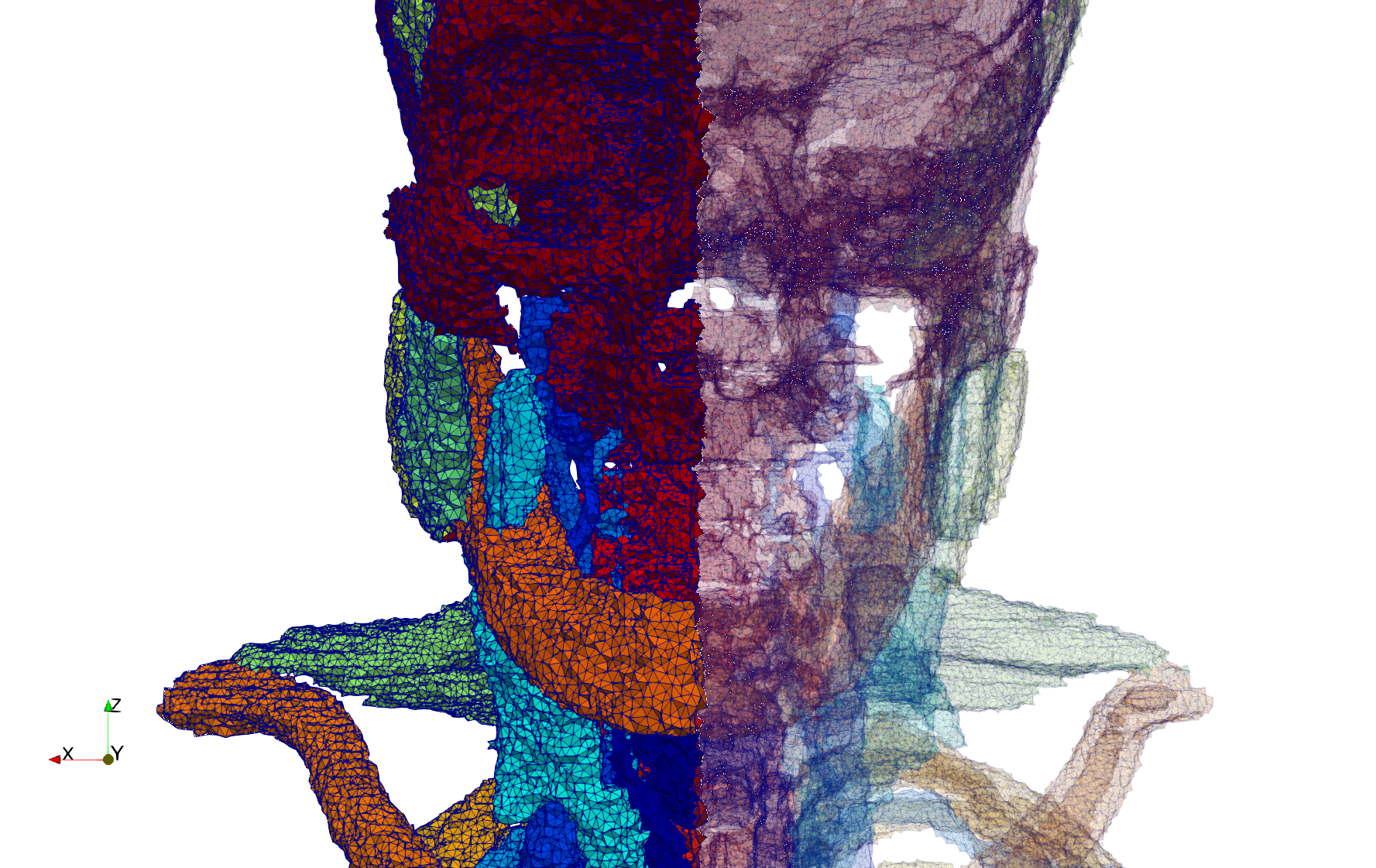
Head-Neck
- Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = default: 205,416 tetrahedra
- Uniform with Delta = 1.5: 770,853 tetrahedra
Commands to generate meshes :
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --output ./Head-Neck,d=2.49023.vtk
Uniform with Delta = 1.5 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
Brain-With-Tumor-Case17
- Input image : Dimensions (448x512x176) with spacing (0.488281x0.488281x1)
- Uniform with Delta = default: 222,540 tetrahedra
- Graded with Delta = default: 94,383 tetrahedra
Commands to generate meshes :
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --output ./Brain-With-Tumor-Case17,d=1.76001.vtk
Graded with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --volume-grading --output ./Brain-With-Tumor-Case17,d=1.76001,graded.vtk
Knee-Char
- Input image : Dimensions (512x512x119) with spacing (0.27734x0.27734x1)
- Uniform with Delta = default case: 386,869 tetrahedra
- Graded with Delta = default case: 274,309 tetrahedra
Commands to generate meshes :
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --output ./Knee-Char,d=1.19.vtk
Graded with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --volume-grading --output ./Knee-Char,d=1.19,graded.vtk
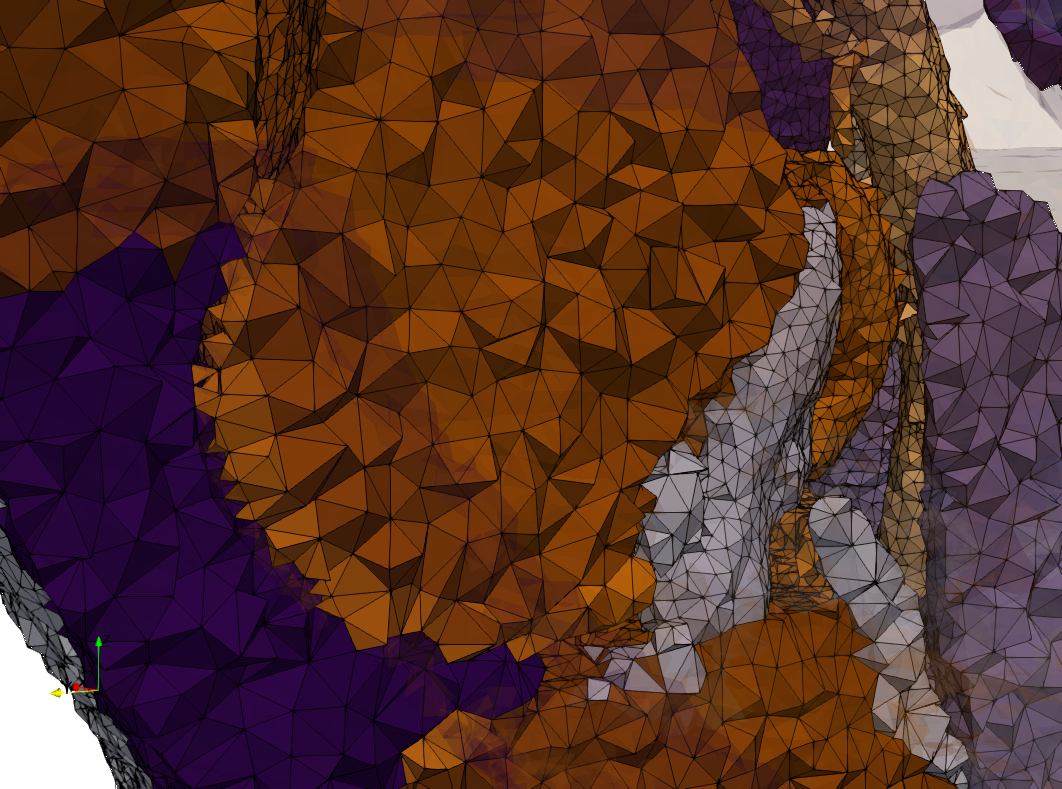
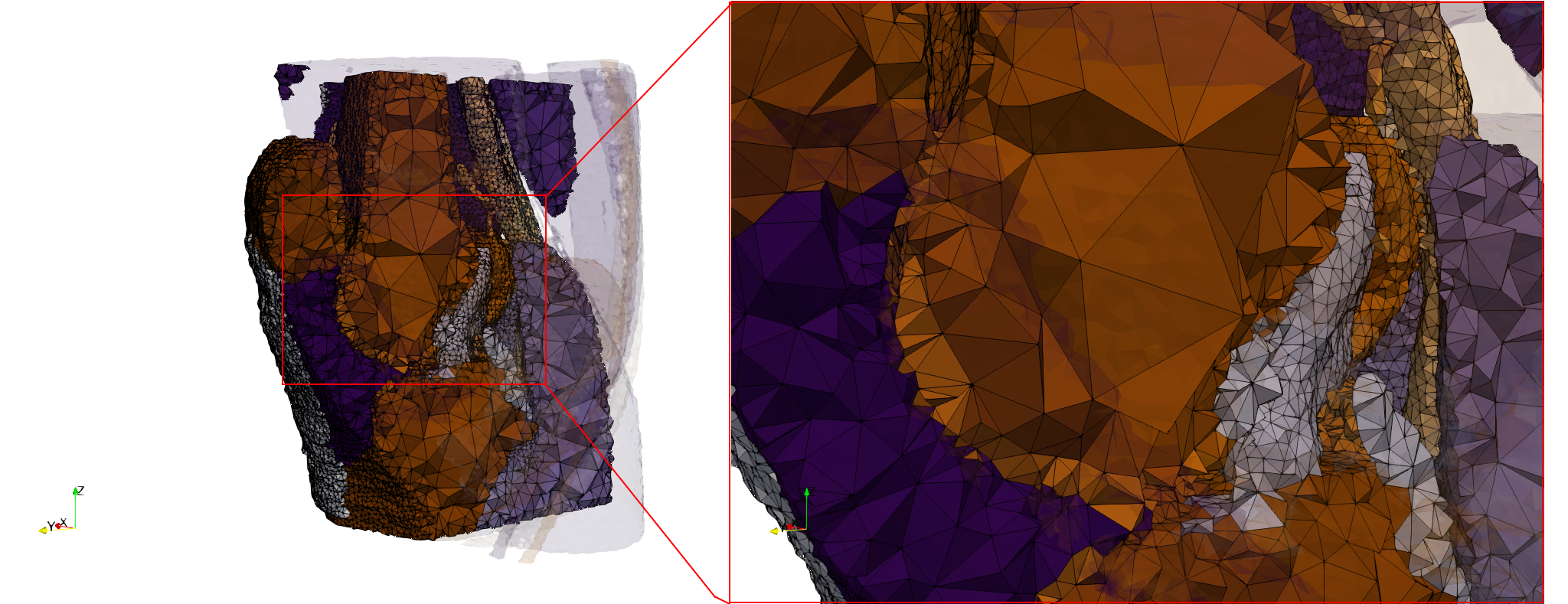
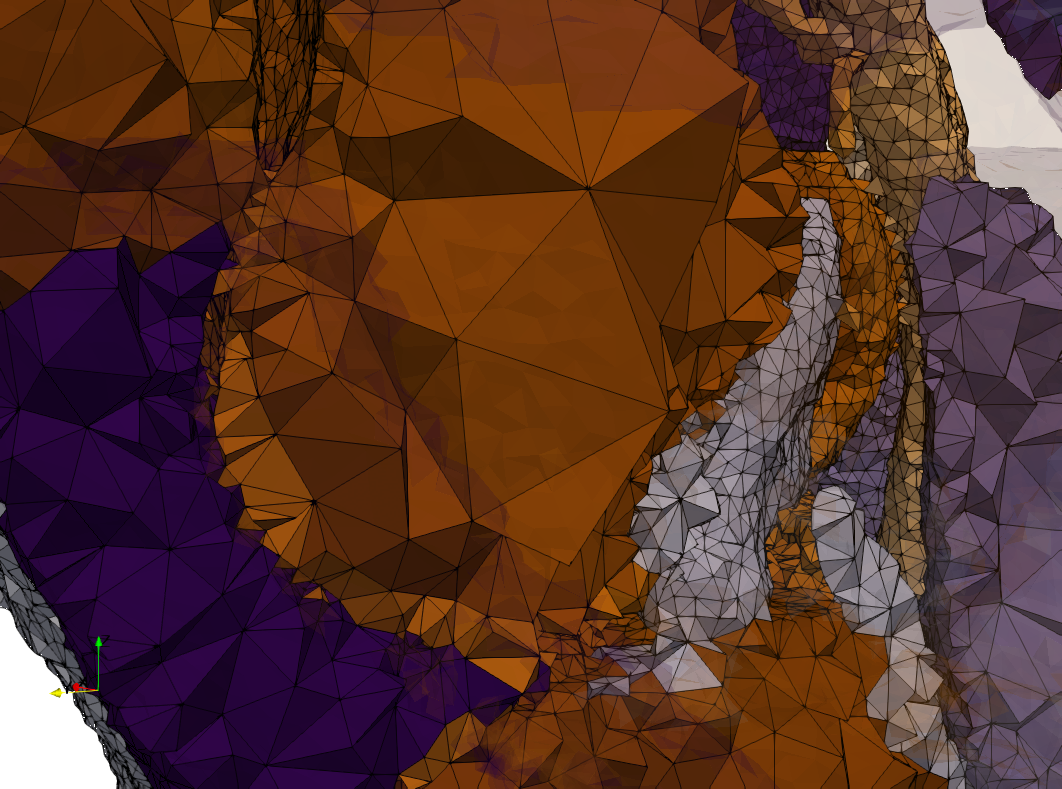
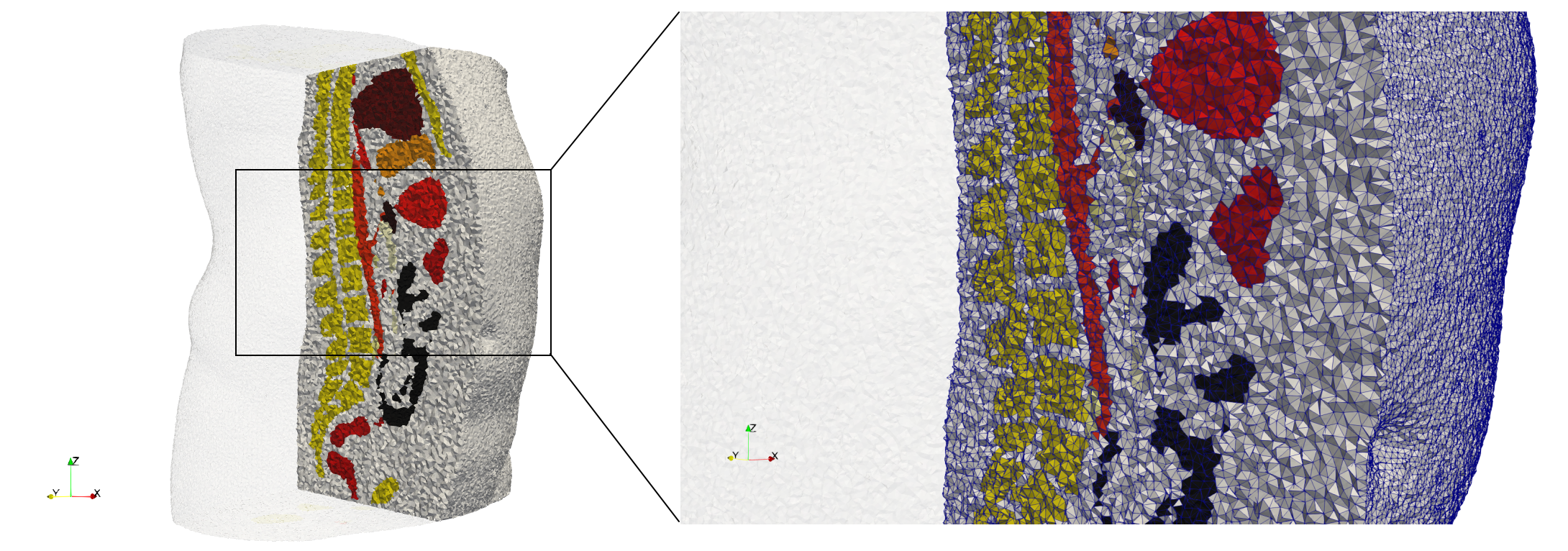
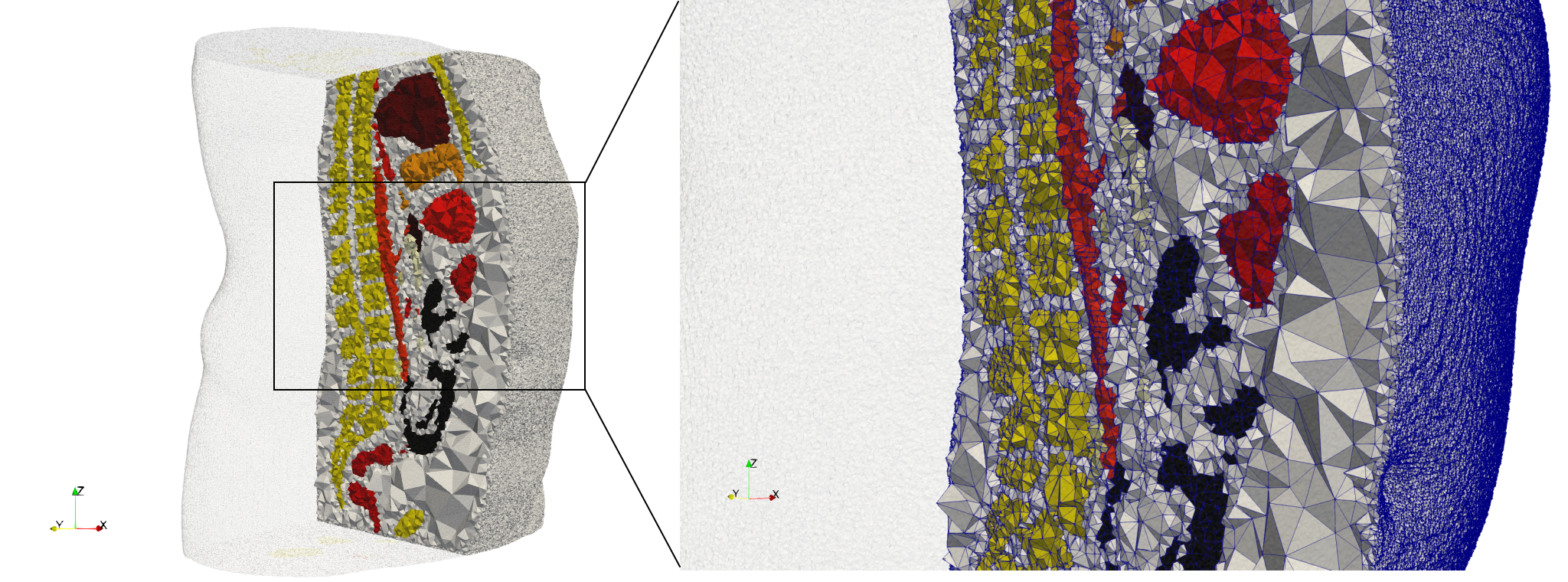
Ircad2
- Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = 2 case: 5,031,442 tetrahedra
- Graded with Delta = 1 case: 6,072,751 tetrahedra
Commands to generate meshes :
Uniform with Delta = 2 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 2 --threads 4 --output ./Ircad2,d=2.vtk
Graded with Delta = 1 : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 4 --volume-grading --output ./Ircad2,d=1,graded.vtk