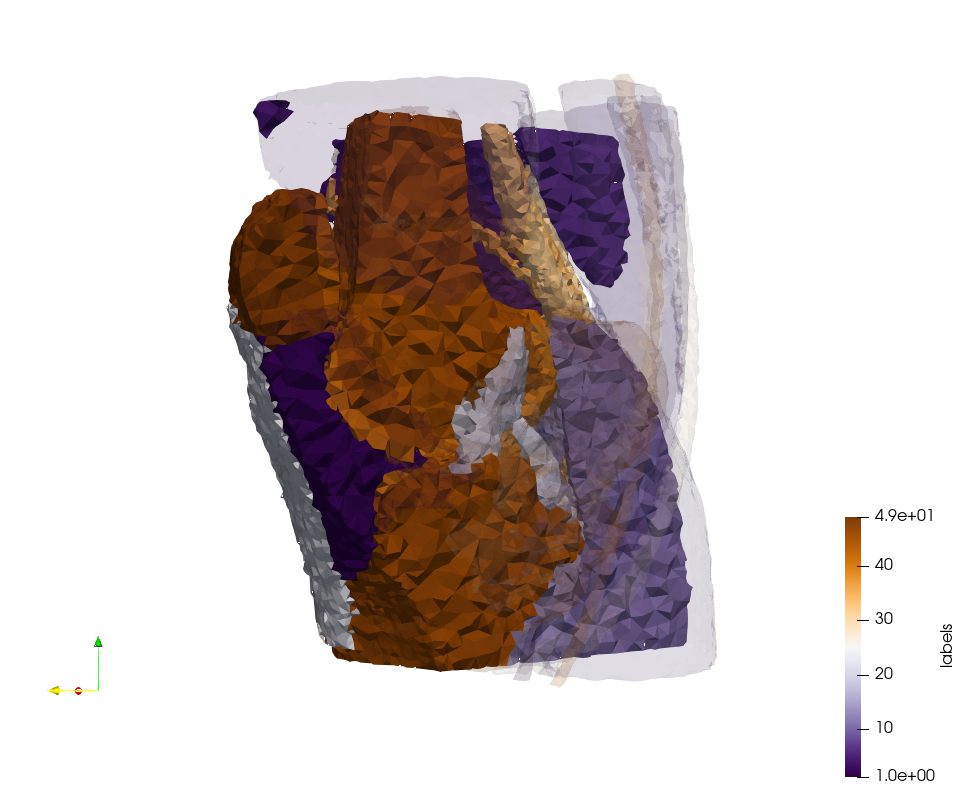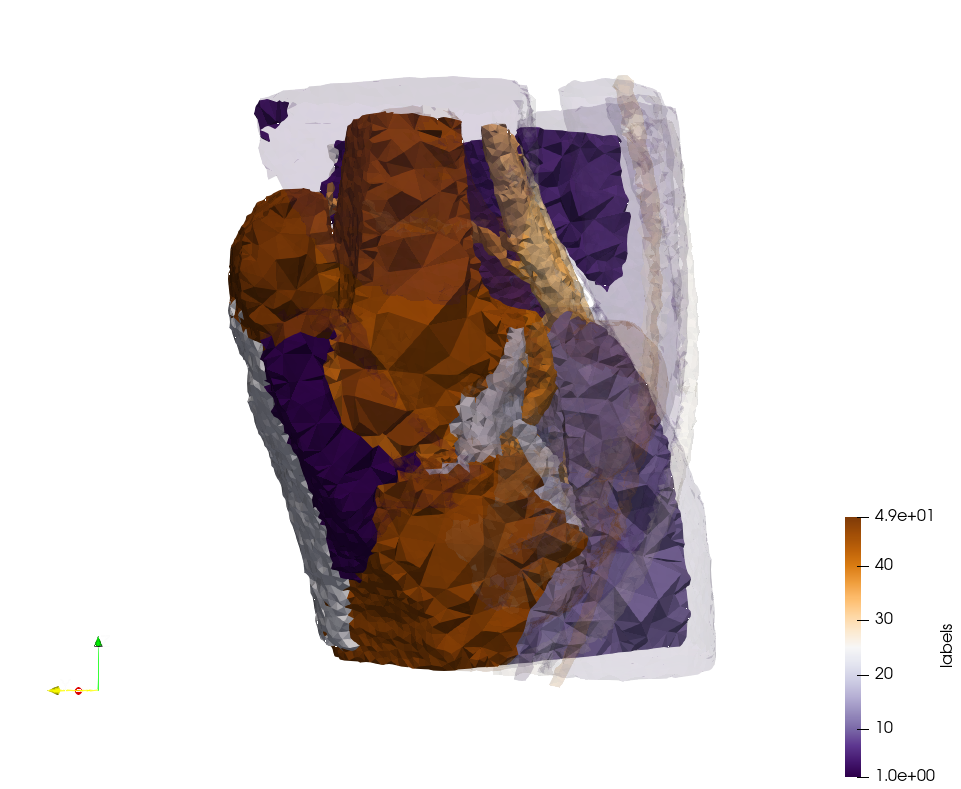Difference between revisions of "Medical Imaging Example Meshes"
From crtc.cs.odu.edu
Spyridon97 (talk | contribs) |
Spyridon97 (talk | contribs) |
||
| Line 29: | Line 29: | ||
Generated mesh with delta = 2, [https://odu.box.com/s/7kgv263qvebk55077nny4l2n7ienpahf Output Mesh] | Generated mesh with delta = 2, [https://odu.box.com/s/7kgv263qvebk55077nny4l2n7ienpahf Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Head-Neck.mha --delta 2 --output ./Head-Neck,d=2.vtk</pre> |
[[File:Head-Neck,d=2.png|500px|Head-Neck,d=2.png]] | [[File:Head-Neck,d=2.png|500px|Head-Neck,d=2.png]] | ||
Generated mesh with delta = 1.5, [https://odu.box.com/s/sfjb83whac8rhn1ykuuxmektprxtci78 Output Mesh] | Generated mesh with delta = 1.5, [https://odu.box.com/s/sfjb83whac8rhn1ykuuxmektprxtci78 Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk</pre> |
[[File:Head-Neck,d=1.5.png|500px|Head-Neck,d=1.5.png]] | [[File:Head-Neck,d=1.5.png|500px|Head-Neck,d=1.5.png]] | ||
| Line 42: | Line 42: | ||
Generated mesh with delta = 1,5, [https://odu.box.com/s/wfeqw6c65si0kync8yi20cychyqxmpcs Output Mesh] | Generated mesh with delta = 1,5, [https://odu.box.com/s/wfeqw6c65si0kync8yi20cychyqxmpcs Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk</pre> |
[[File:Brain-With-Tumor-Case17,d=1.5.png|500px|Brain-With-Tumor-Case17,d=1.5.png]] | [[File:Brain-With-Tumor-Case17,d=1.5.png|500px|Brain-With-Tumor-Case17,d=1.5.png]] | ||
Generated mesh with delta = 1.5 and volume-grading, [https://odu.box.com/s/83pl6vgzabzclsbqyzujmtqjbfckil2s Output Mesh] | Generated mesh with delta = 1.5 and volume-grading, [https://odu.box.com/s/83pl6vgzabzclsbqyzujmtqjbfckil2s Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk</pre> |
[[File:Brain-With-Tumor-Case17,d=1.5,grading.png|500px|Brain-With-Tumor-Case17,d=1.5,grading.png]] | [[File:Brain-With-Tumor-Case17,d=1.5,grading.png|500px|Brain-With-Tumor-Case17,d=1.5,grading.png]] | ||
| Line 55: | Line 55: | ||
Generated mesh with delta = 1,5, [https://odu.box.com/s/udhiz0u5bewtqi9z07owiejno9gjzb1j Output Mesh] | Generated mesh with delta = 1,5, [https://odu.box.com/s/udhiz0u5bewtqi9z07owiejno9gjzb1j Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk</pre> |
[[File:Knee-Char,d=1.5.png|500px|Knee-Char,d=1.5.png]] | [[File:Knee-Char,d=1.5.png|500px|Knee-Char,d=1.5.png]] | ||
Generated mesh with delta = 1.5 and volume-grading, [https://odu.box.com/s/tmmv1kjb925zclnshegypovxmaxablcs Output Mesh] | Generated mesh with delta = 1.5 and volume-grading, [https://odu.box.com/s/tmmv1kjb925zclnshegypovxmaxablcs Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk</pre> |
[[File:Knee-Char,d=1.5,grading.png|500px|Knee-Char,d=1.5,grading.png]] | [[File:Knee-Char,d=1.5,grading.png|500px|Knee-Char,d=1.5,grading.png]] | ||
| Line 68: | Line 68: | ||
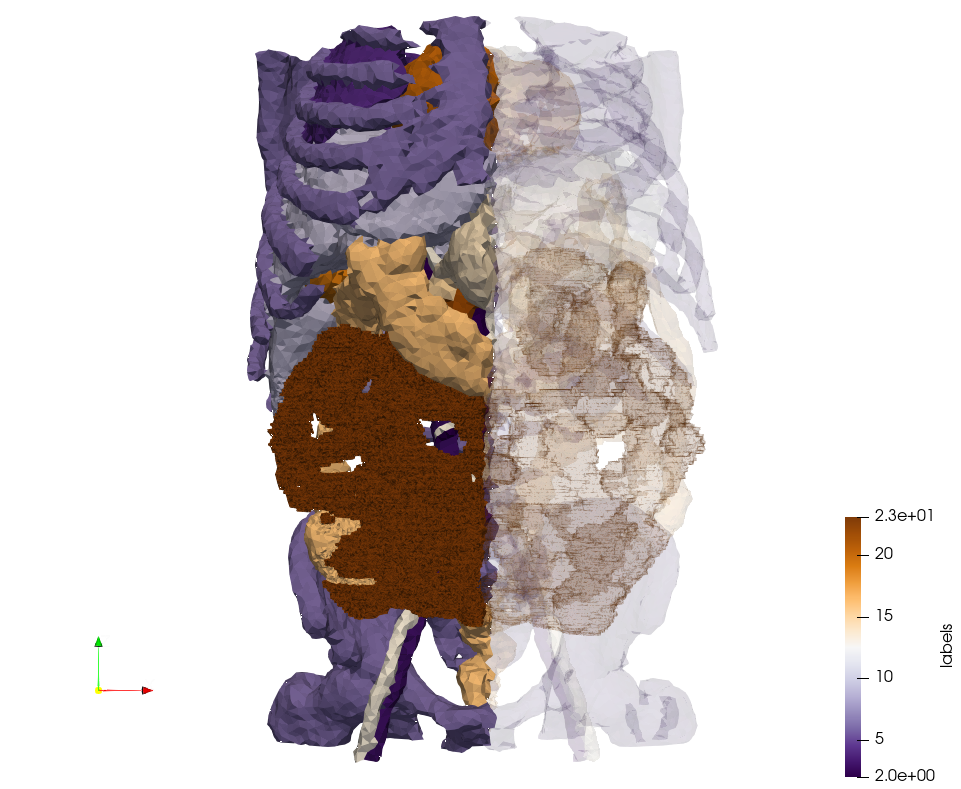
Generated mesh with delta = 5, [https://odu.box.com/s/ddja85vt80osbn76v7pih82wh0kiuqqp Output Mesh] | Generated mesh with delta = 5, [https://odu.box.com/s/ddja85vt80osbn76v7pih82wh0kiuqqp Output Mesh] | ||
| − | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./ | + | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk</pre> |
[[File:Ircad2-Removed-Tissues,d=5.png|500px|Ircad2-Removed-Tissues,d=5.png]] | [[File:Ircad2-Removed-Tissues,d=5.png|500px|Ircad2-Removed-Tissues,d=5.png]] | ||
Revision as of 16:57, 26 February 2020
The directory containing the input data is Medical_Imaging_Data.
Head-Neck 2
- Input distribution size : 1,000,000 cells
- Uniform case: 768,033 tetrahedra
- Adapted case: 264,762 tetrahedra
Commands to generate meshes :
Uniform : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./CNF_Data/3D/CFF_14052019/GPDMMS13_140519.nrrd --image-segmentation --linear-interpolation --output ./GPDMMS13_140519,d=1,uniform.vtk
Adapted : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./CNF_Data/3D/CFF_14052019/GPDMMS13_140519.nrrd --cnf --weight-limit 0.05 --min-edge 1 --output ./GPDMMS13_140519,d=5,wl=0.05,me=1.vtk
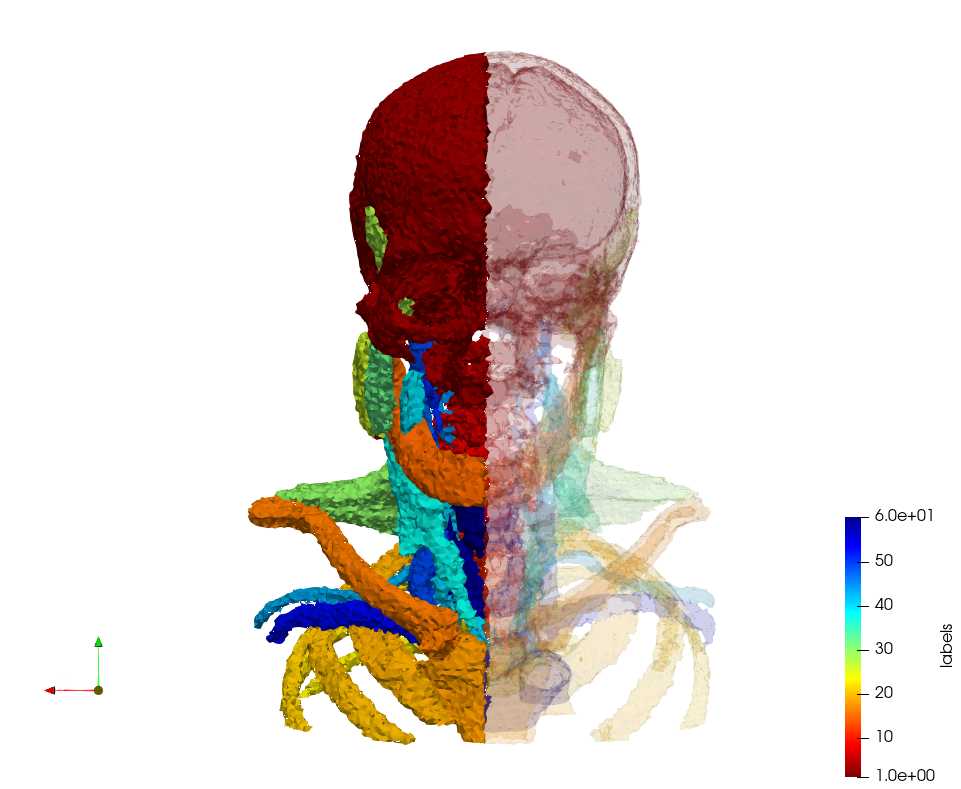
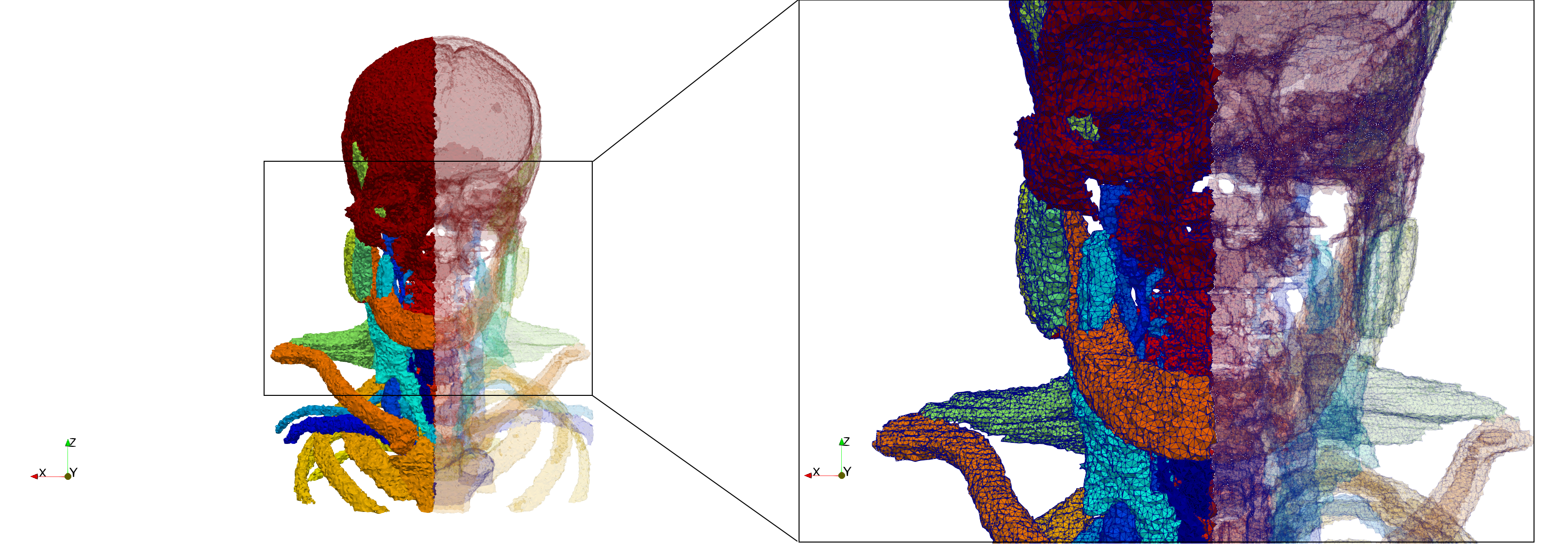
Head-Neck
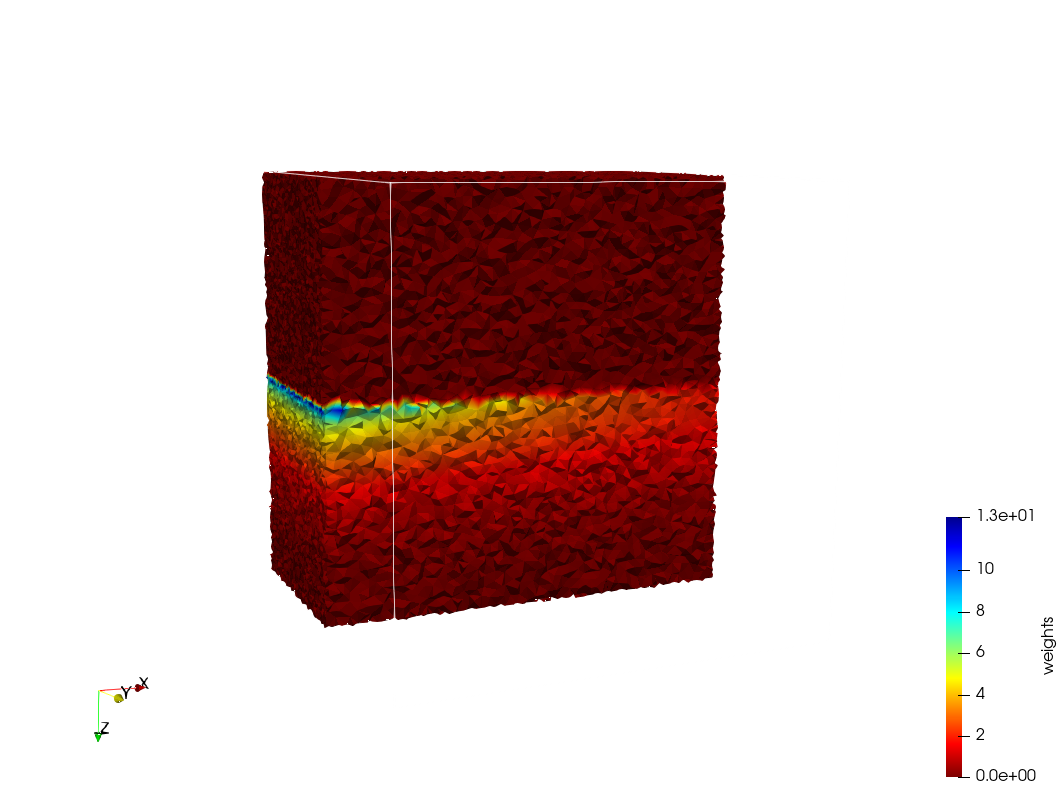
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Head-Neck.mha --delta 2 --output ./Head-Neck,d=2.vtk
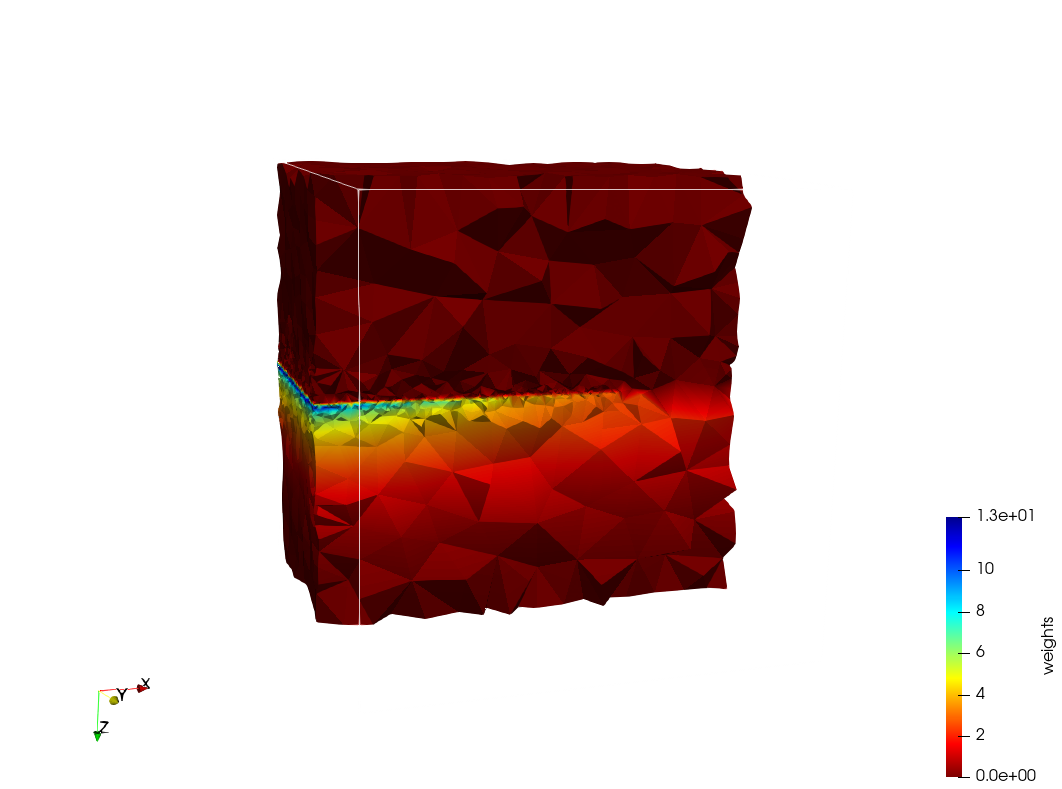
Generated mesh with delta = 1.5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
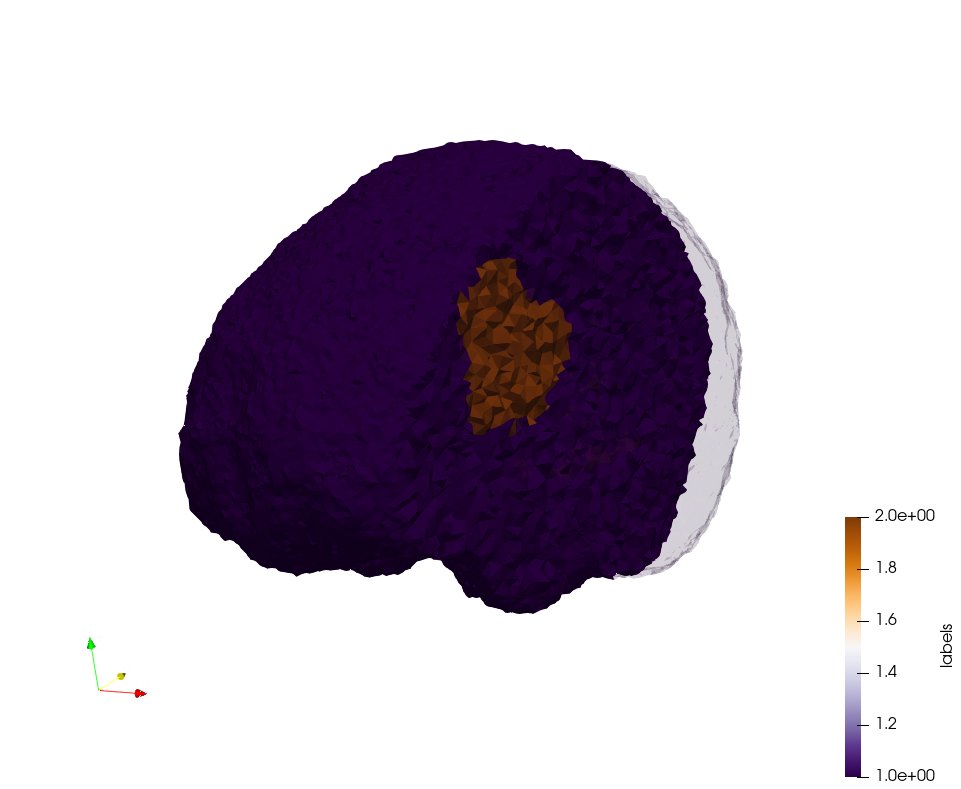
Brain-With-Tumor-Case17
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk
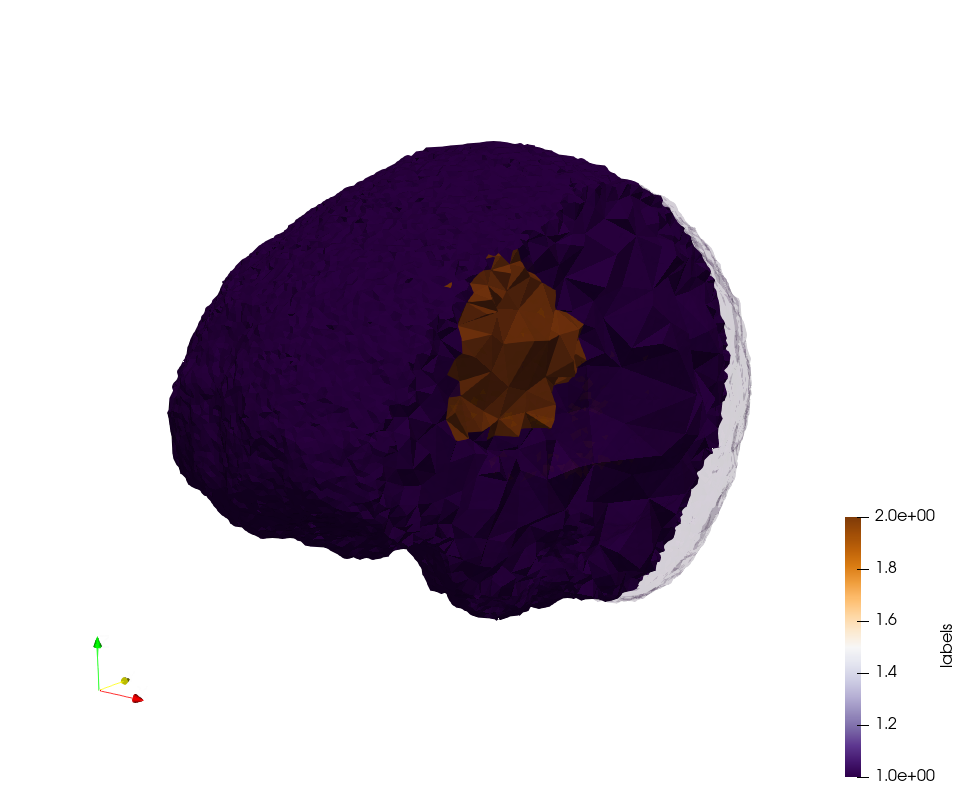
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk
Knee-Char
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk
Ircad2-Removed-Tissues
Generated mesh with delta = 5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk