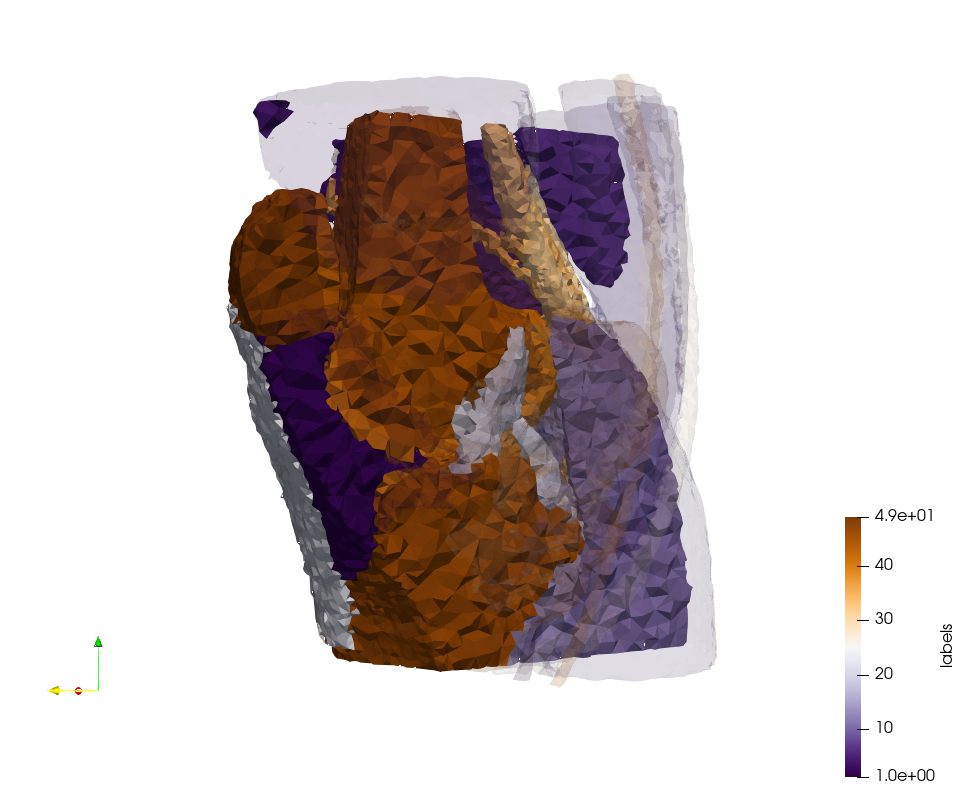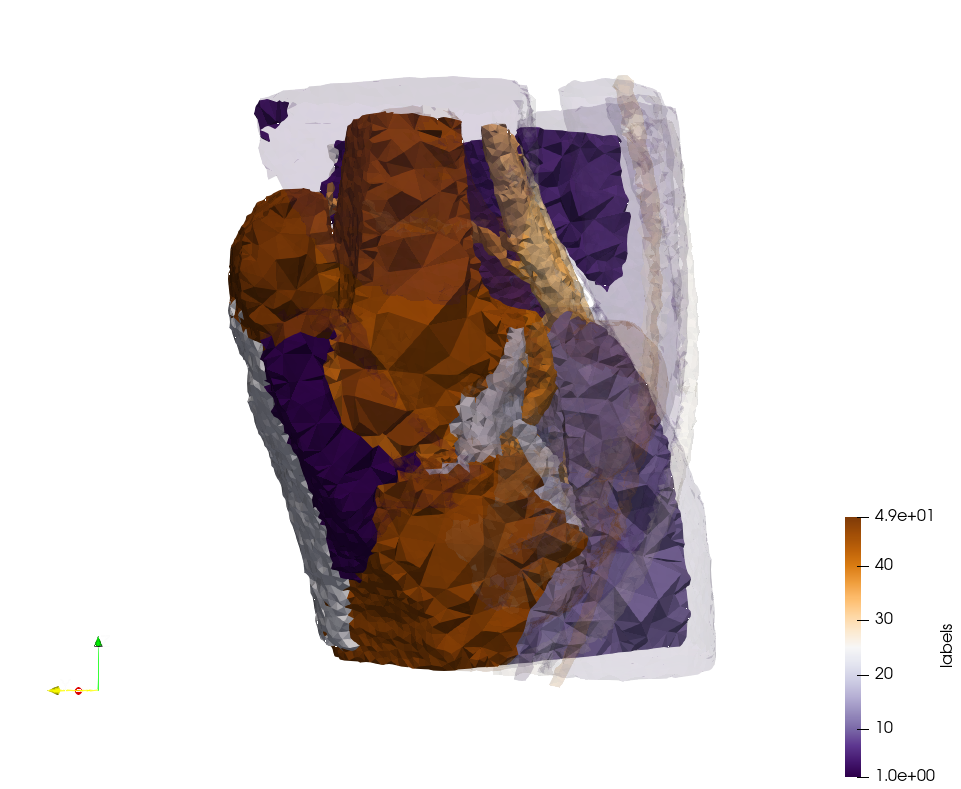Difference between revisions of "Medical Imaging Example Meshes"
From crtc.cs.odu.edu
Spyridon97 (talk | contribs) |
Spyridon97 (talk | contribs) |
||
| Line 1: | Line 1: | ||
| + | The directory containing the input data is [https://odu.box.com/s/wd0i18giti2zo330y19xhkqu8wjzgf7j Medical_Imaging_Data]. | ||
| + | |||
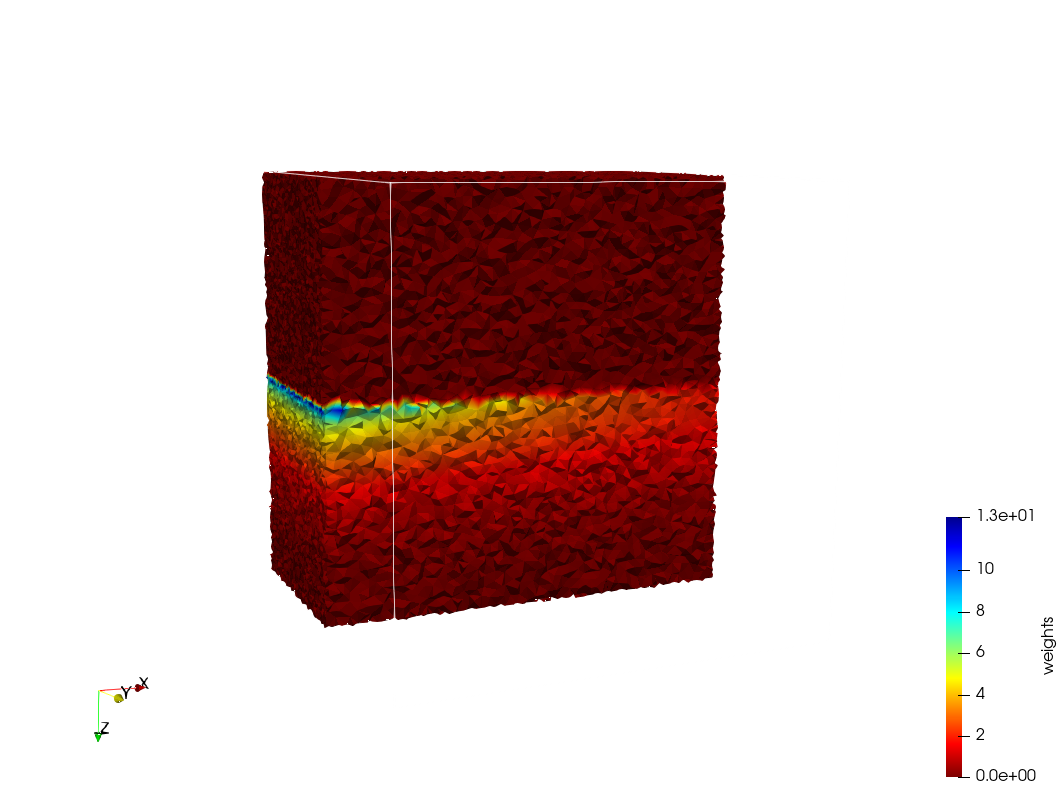
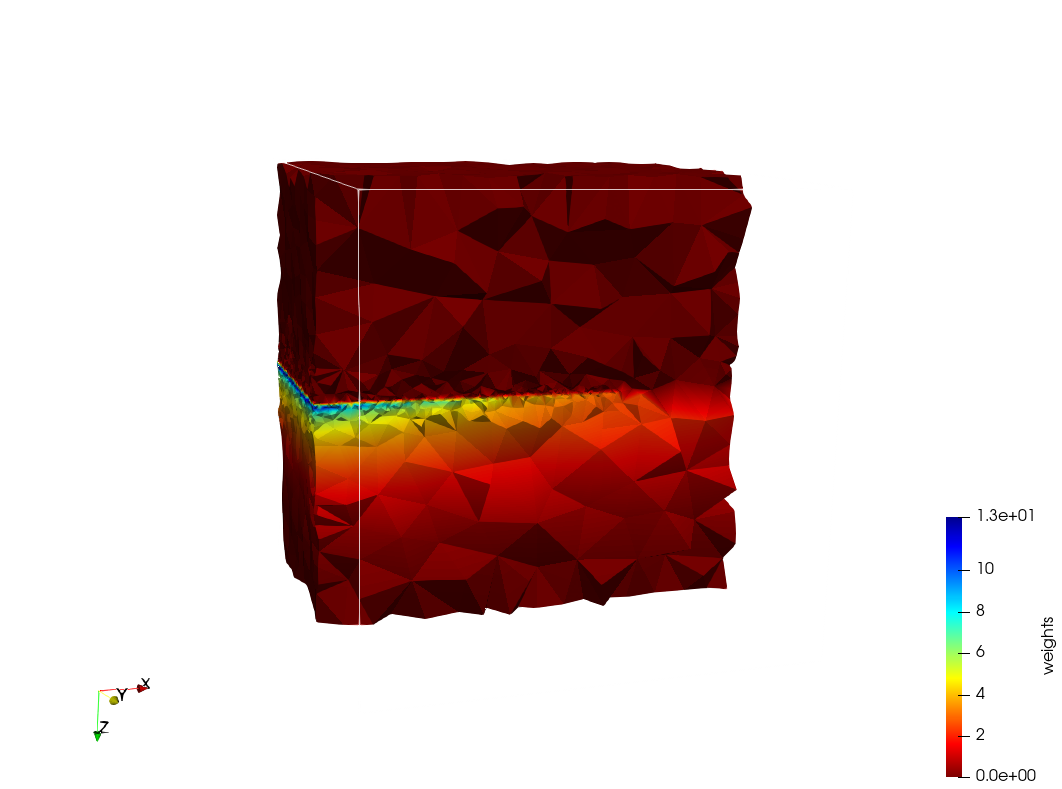
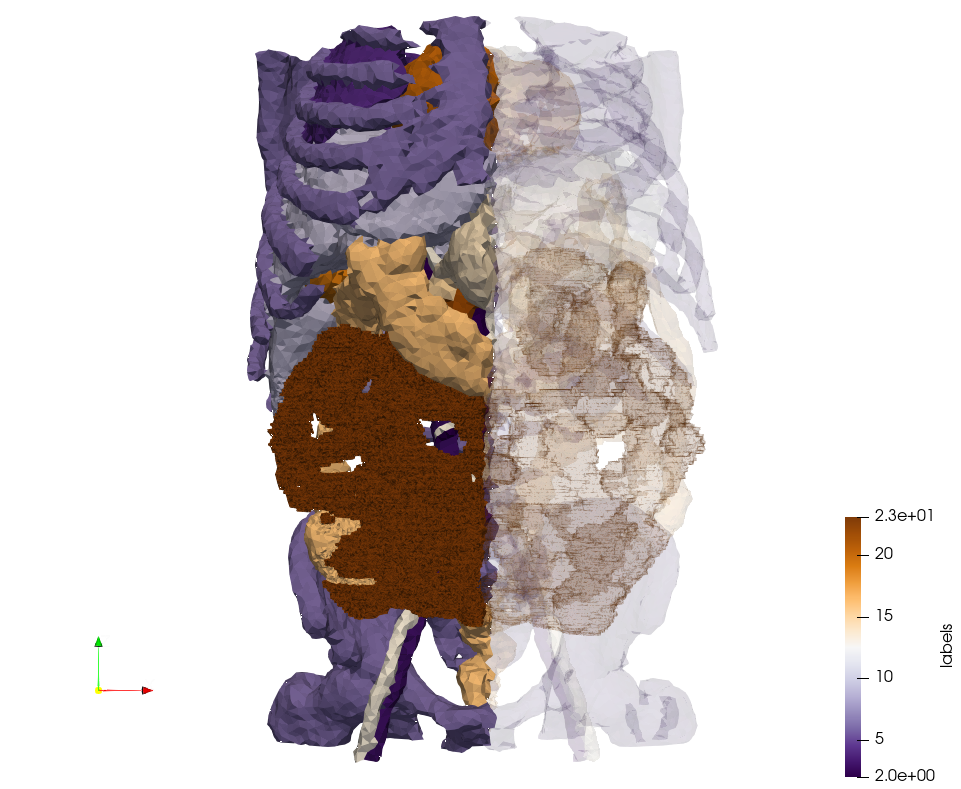
| + | =Head-Neck 2 = | ||
| + | * Input distribution size : 1,000,000 cells | ||
| + | * Uniform case: 768,033 tetrahedra | ||
| + | * Adapted case: 264,762 tetrahedra | ||
| + | |||
| + | <gallery mode="packed" heights=350px> | ||
| + | File:GPDMMS13_140519,d=1,uniform.png | ||
| + | File:GPDMMS13_140519,d=5,wl=0.05,me=1.png | ||
| + | </gallery> | ||
| + | |||
| + | Commands to generate meshes : | ||
| + | |||
| + | '''Uniform :''' [https://odu.box.com/s/h48toji5cii2rk6xkmofptnw4nntt8zk Output Mesh] | ||
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./CNF_Data/3D/CFF_14052019/GPDMMS13_140519.nrrd --image-segmentation --linear-interpolation --output ./GPDMMS13_140519,d=1,uniform.vtk | ||
| + | </pre> | ||
| + | |||
| + | '''Adapted :''' [https://odu.box.com/s/bcg7aw4lv9rvp4531va7pqg7j2os7tny Output Mesh] | ||
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./CNF_Data/3D/CFF_14052019/GPDMMS13_140519.nrrd --cnf --weight-limit 0.05 --min-edge 1 --output ./GPDMMS13_140519,d=5,wl=0.05,me=1.vtk | ||
| + | </pre> | ||
| + | |||
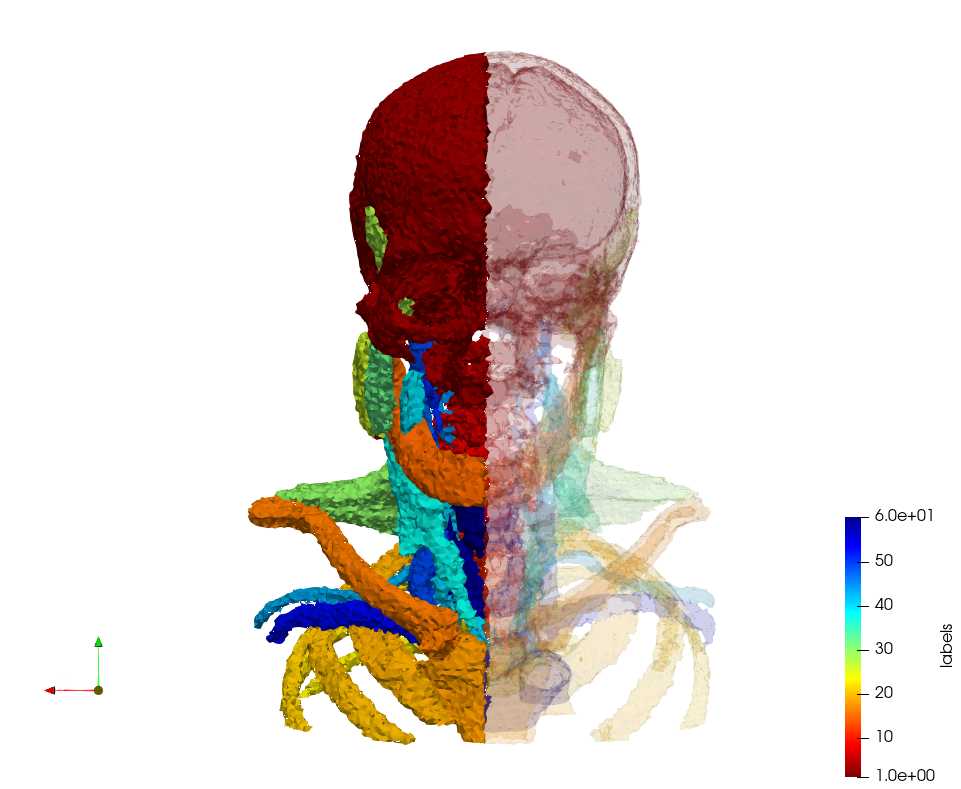
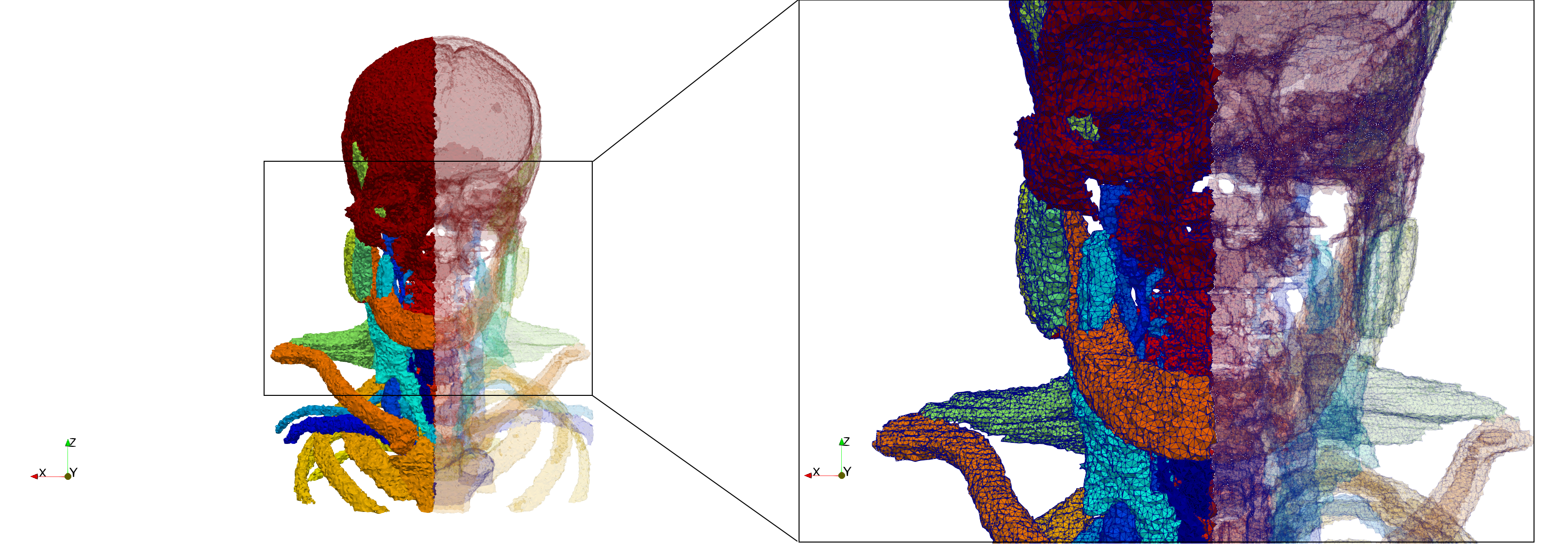
=Head-Neck= | =Head-Neck= | ||
[https://odu.box.com/s/09l553pqu319jdj3isam6dmukfy67ouq Input Image] | [https://odu.box.com/s/09l553pqu319jdj3isam6dmukfy67ouq Input Image] | ||
Revision as of 16:52, 26 February 2020
The directory containing the input data is Medical_Imaging_Data.
Head-Neck 2
- Input distribution size : 1,000,000 cells
- Uniform case: 768,033 tetrahedra
- Adapted case: 264,762 tetrahedra
Commands to generate meshes :
Uniform : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./CNF_Data/3D/CFF_14052019/GPDMMS13_140519.nrrd --image-segmentation --linear-interpolation --output ./GPDMMS13_140519,d=1,uniform.vtk
Adapted : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./CNF_Data/3D/CFF_14052019/GPDMMS13_140519.nrrd --cnf --weight-limit 0.05 --min-edge 1 --output ./GPDMMS13_140519,d=5,wl=0.05,me=1.vtk
Head-Neck
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 2 --output ./Head-Neck,d=2.vtk
Generated mesh with delta = 1.5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
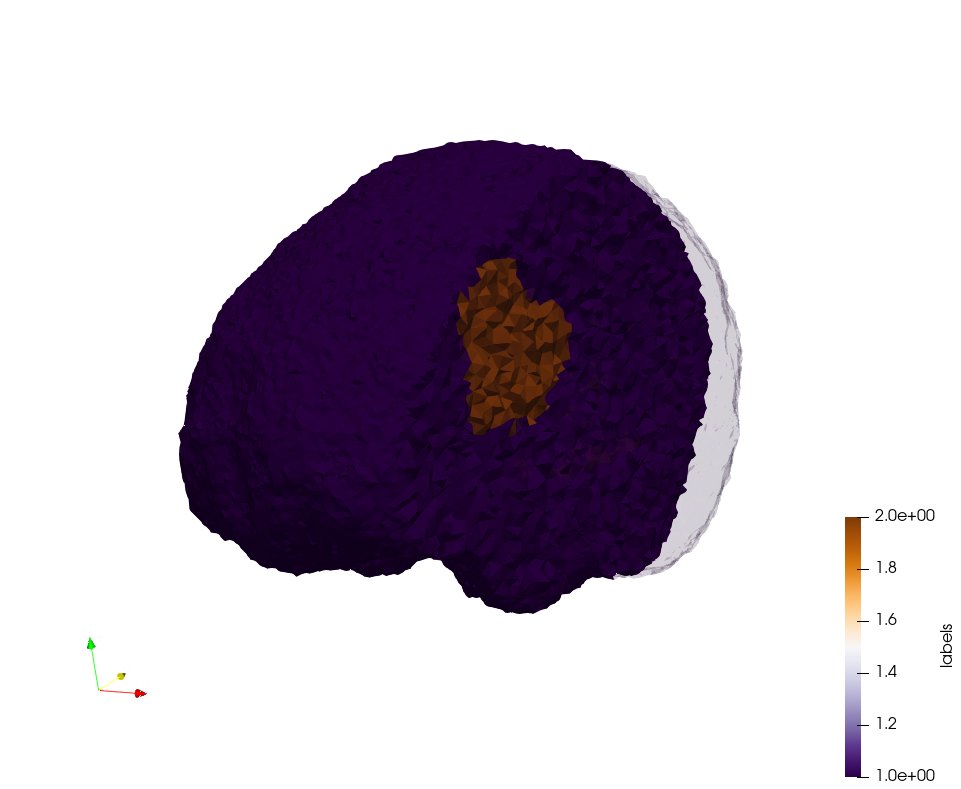
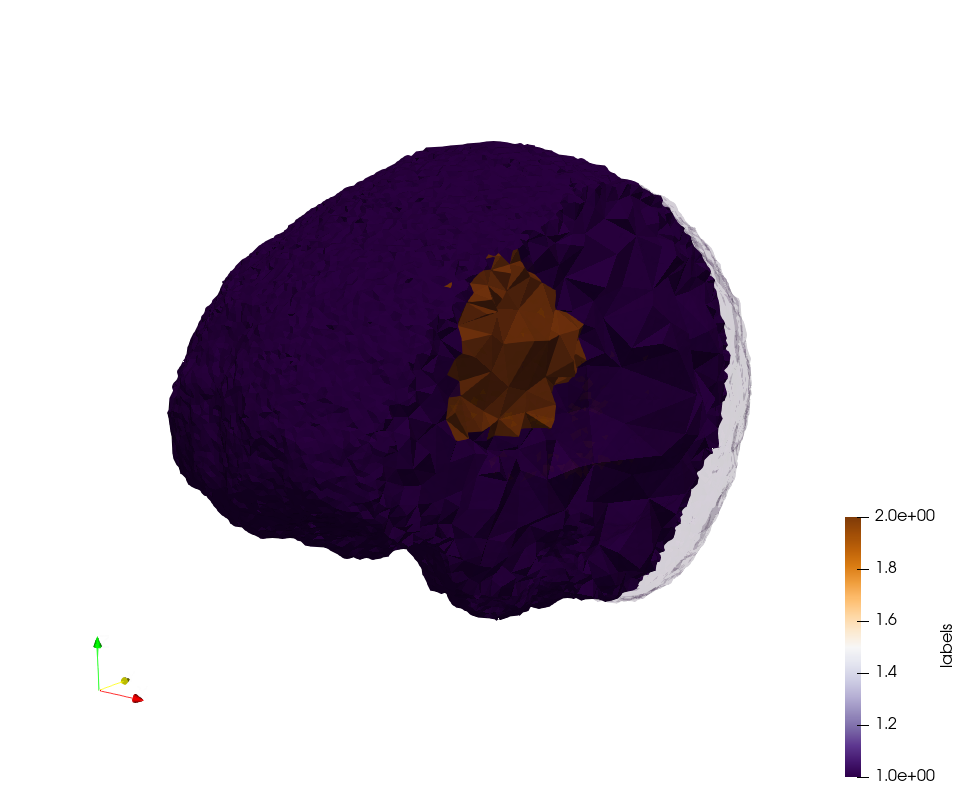
Brain-With-Tumor-Case17
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk
Knee-Char
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk
Ircad2-Removed-Tissues
Generated mesh with delta = 5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk