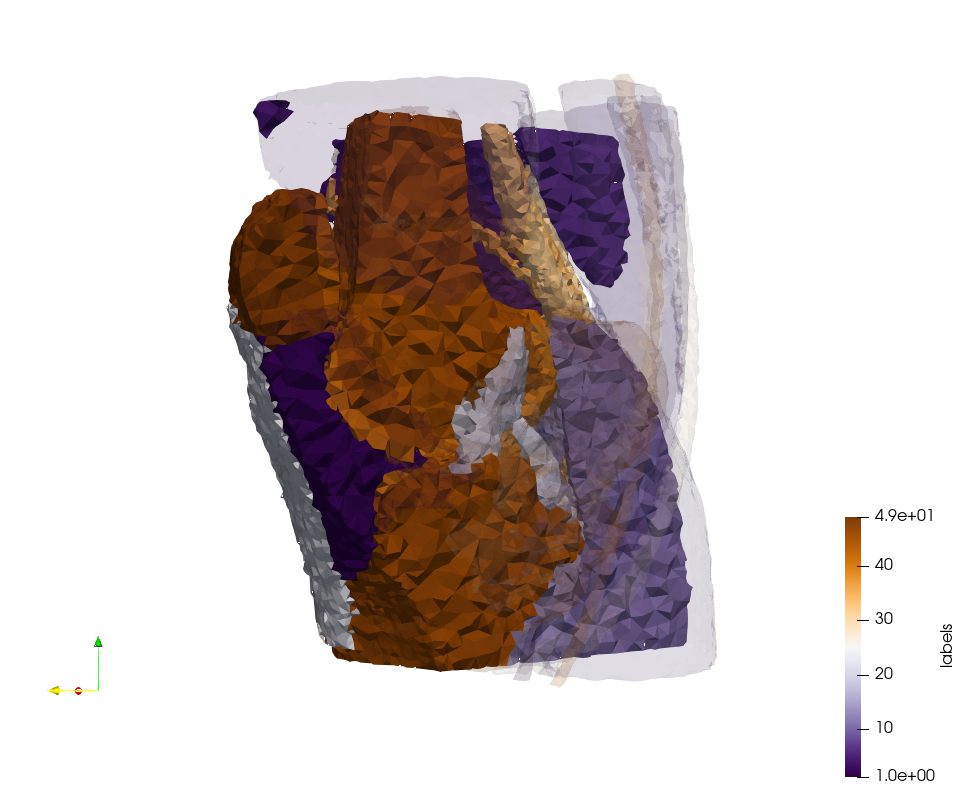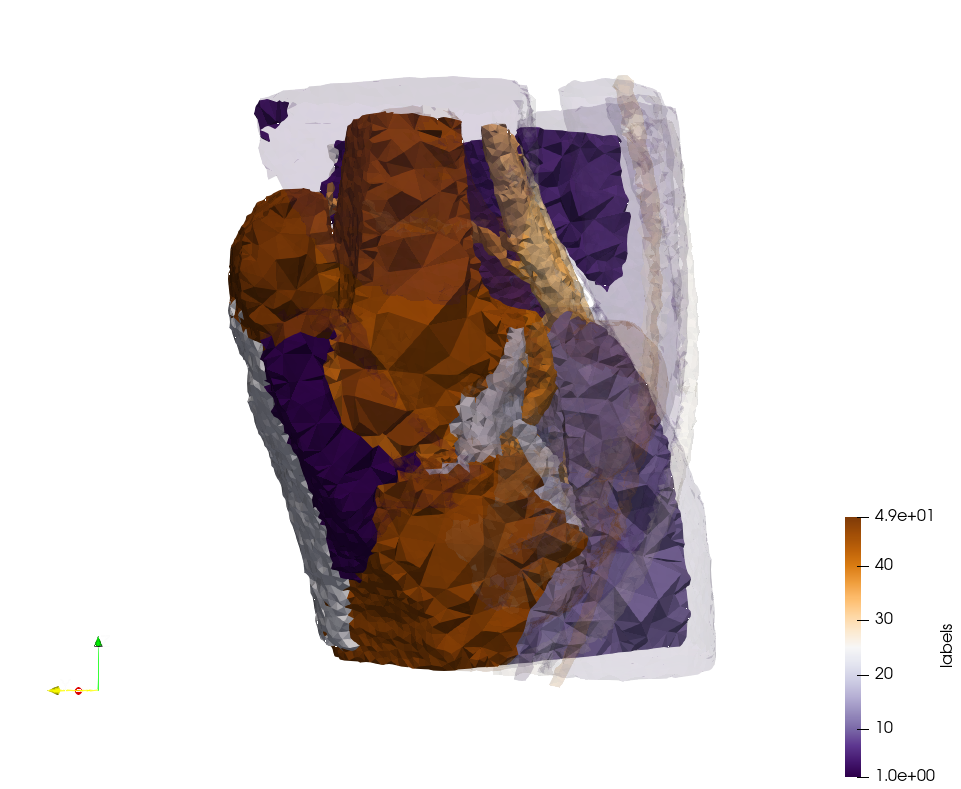Difference between revisions of "Medical Imaging Example Meshes"
From crtc.cs.odu.edu
Spyridon97 (talk | contribs) (→Role of Delta) |
Spyridon97 (talk | contribs) |
||
| Line 66: | Line 66: | ||
[https://odu.box.com/s/01bxjptf4grfuvxj3a9g6frfosyu6dbt Input Image] | [https://odu.box.com/s/01bxjptf4grfuvxj3a9g6frfosyu6dbt Input Image] | ||
| − | Generated mesh with delta = 5, | + | Generated mesh with delta = 5, https://odu.box.com/s/ddja85vt80osbn76v7pih82wh0kiuqqp Output Mesh] |
<pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk</pre> | <pre>docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk</pre> | ||
[[File:Ircad2-Removed-Tissues,d=5.png|500px|Ircad2-Removed-Tissues,d=5.png]] | [[File:Ircad2-Removed-Tissues,d=5.png|500px|Ircad2-Removed-Tissues,d=5.png]] | ||
Revision as of 14:40, 18 February 2020
Contents
Role of Delta
Table of produced number of Tetrahedra for certain values of delta using the image: Brain-With-Tumor-Case17.
The size of the input image is: (448,512,176) Spacing of the input image: (0.488281,0.488281,1)
MinimumPhysicalSize = 176 * 1 = 176
| Delta | Amount of Tetrahedra |
|---|---|
| 2.21 | 7181 |
| 2.34 | 5922 |
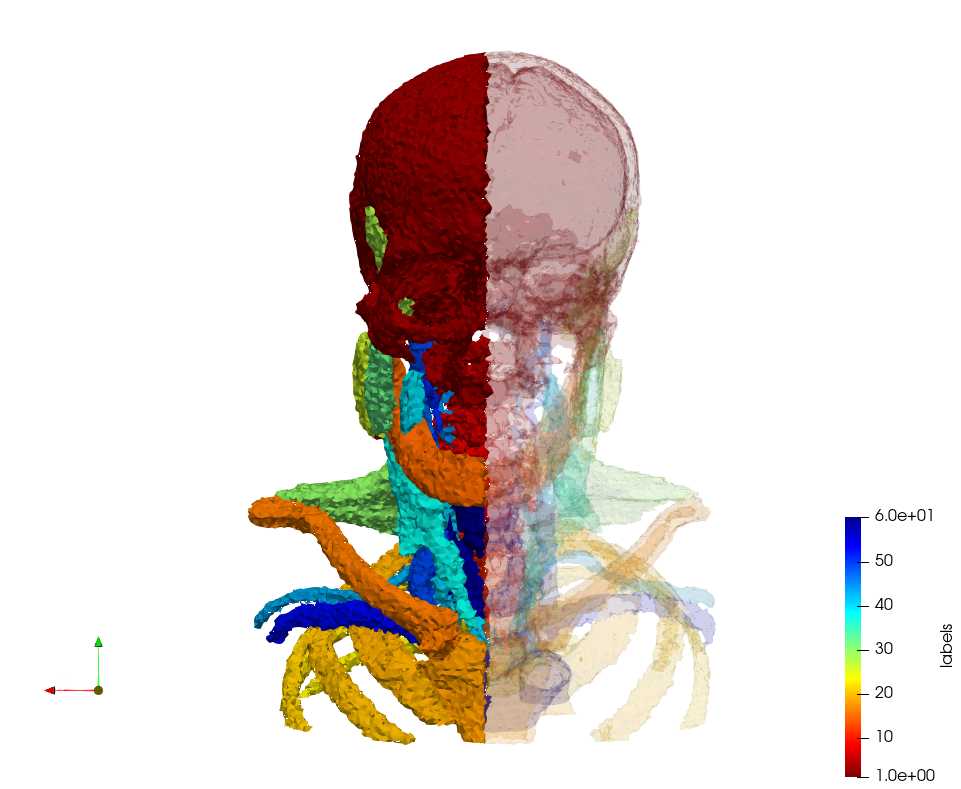
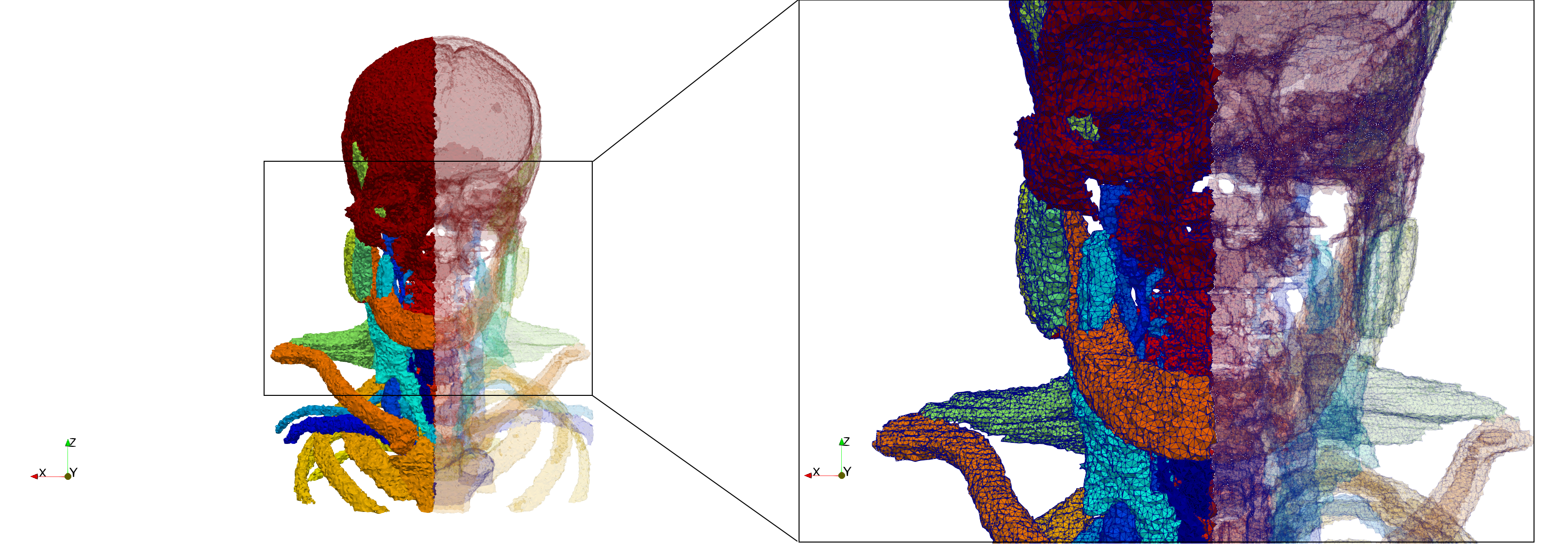
Head-Neck
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 2 --output ./Head-Neck,d=2.vtk
Generated mesh with delta = 1.5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
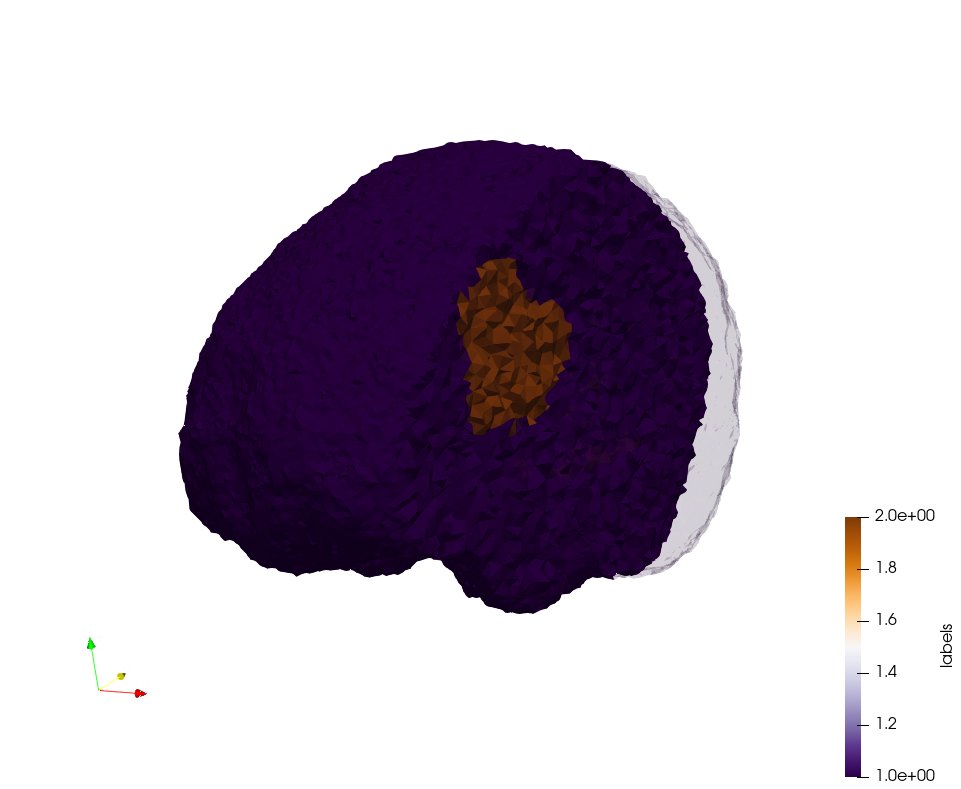
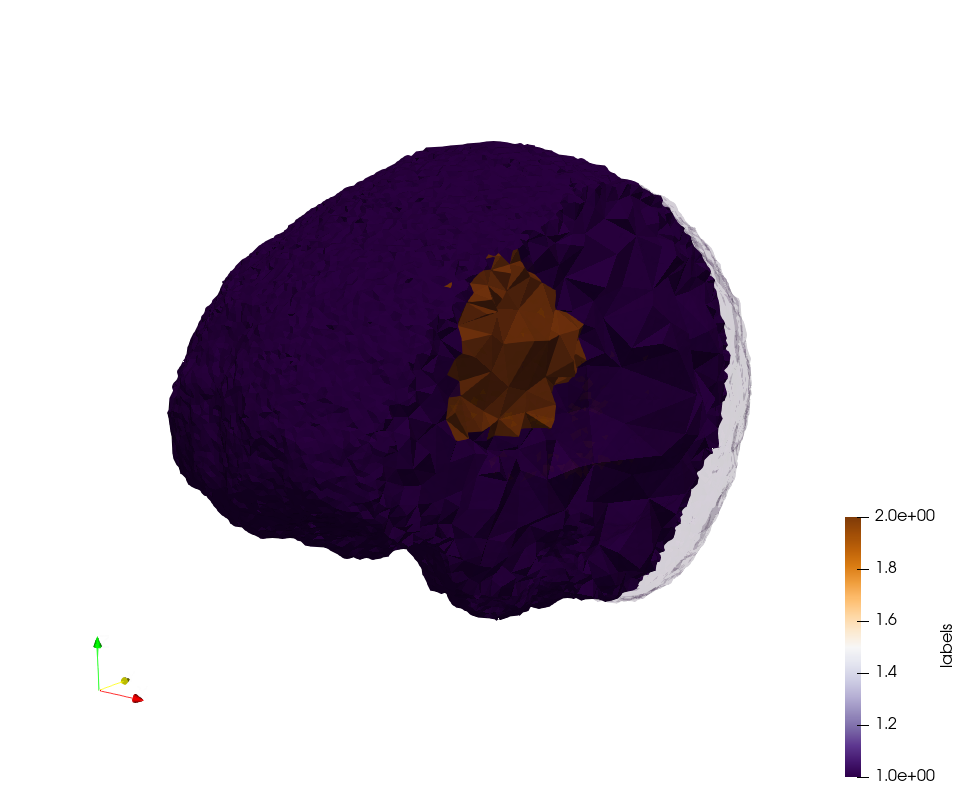
Brain-With-Tumor-Case17
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk
Knee-Char
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk
Big-Brain
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Big-Brain.mha --delta 2 --output ./Big-Brain,d=2.vtk
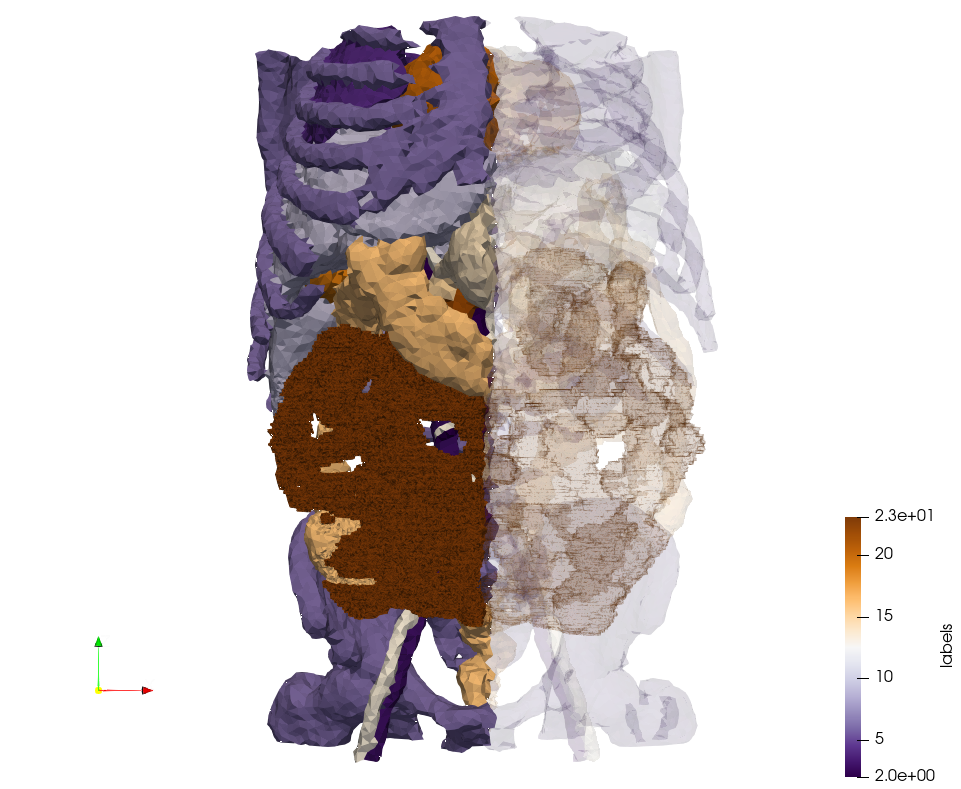
Ircad2-Removed-Tissues
Generated mesh with delta = 5, https://odu.box.com/s/ddja85vt80osbn76v7pih82wh0kiuqqp Output Mesh]
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk