Bioinformatics Example Meshes
Contents
3D Example Meshes
The directory containing the 3D input data is located in the 3D folder of Bioninformatics_Data.
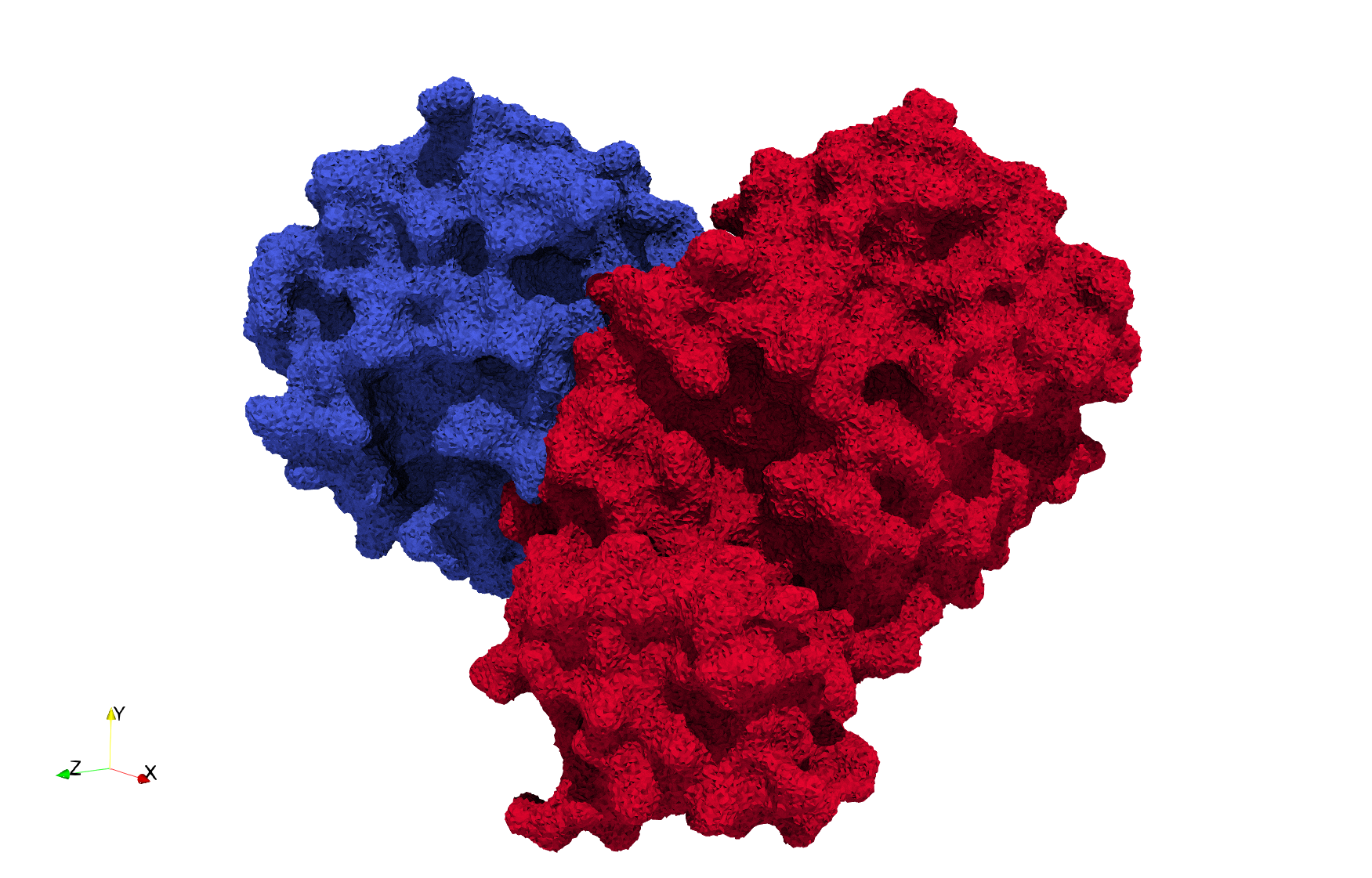
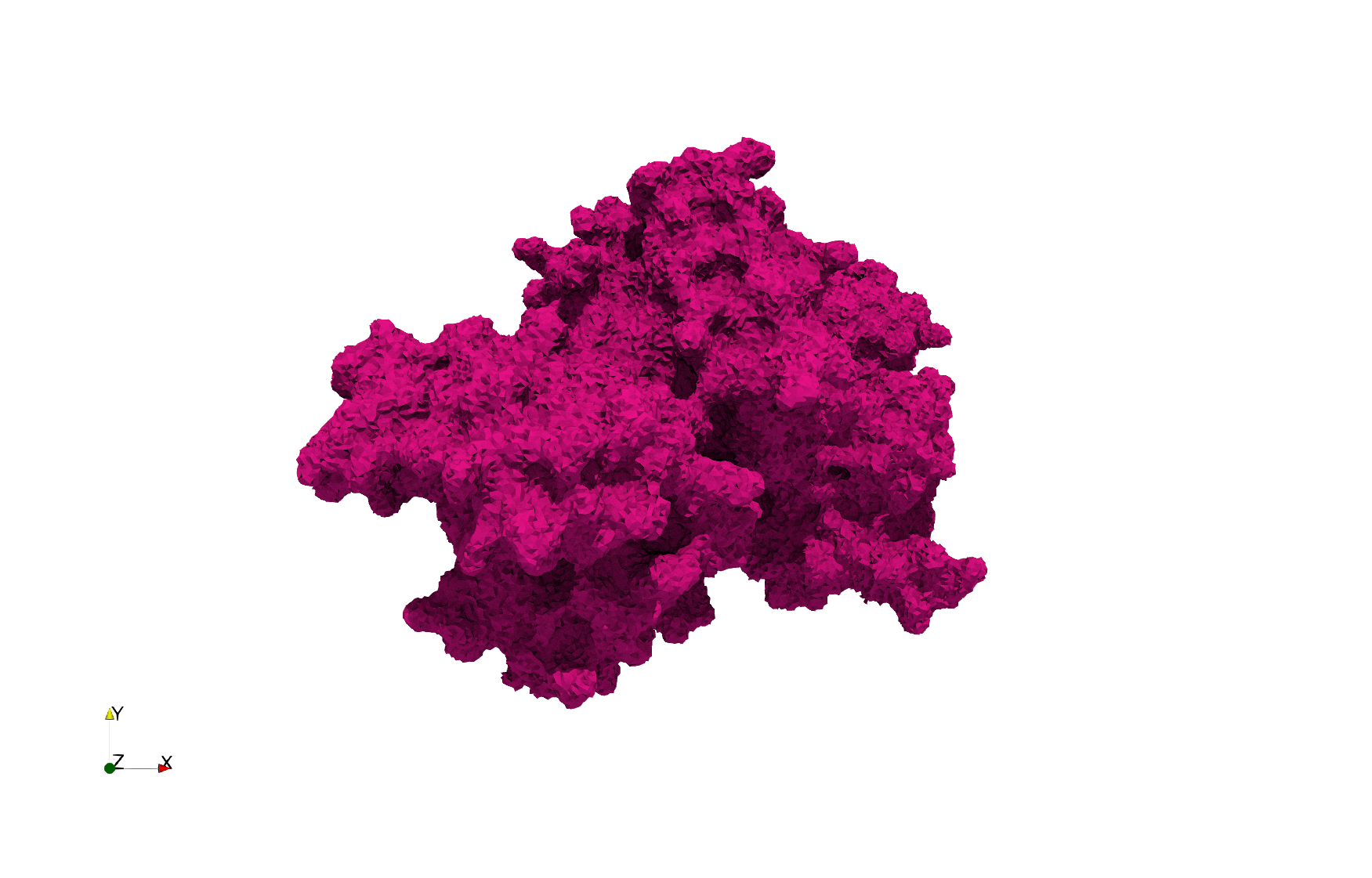
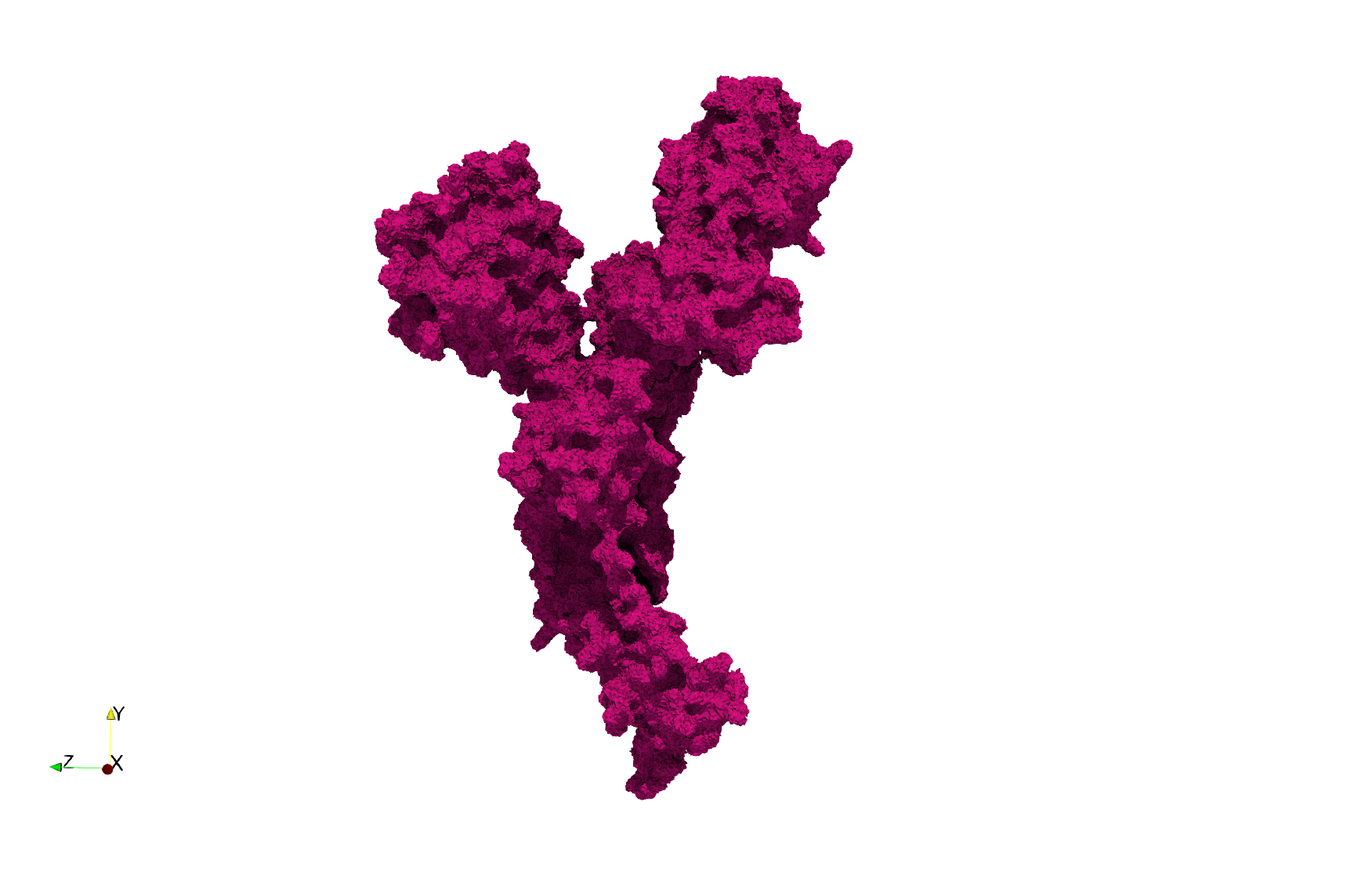
COVID-19-Main-Protease-6y2e
- Input Image
- Input image : Dimensions (284x303x344) with spacing (0.2226907x0.2226907x0.2226907)
- Uniform with Delta = 0.3: 2,509,202 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.3: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Main-Protease-6y2e.nrrd --delta 0.3 --output ./COVID-19-Main-Protease-6y2e,d=0.3.vtk
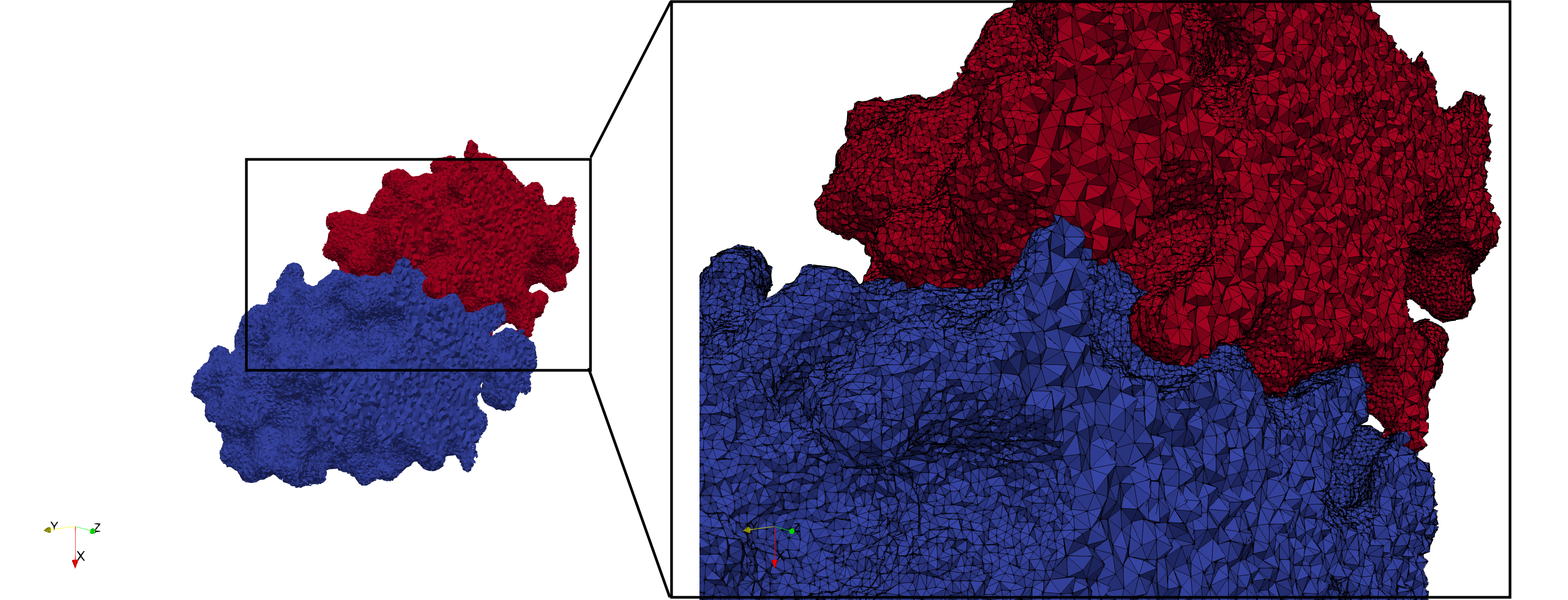
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 Main Protease. Biological assembly data was retrieved from the molecule 6y2e, Protein Data Bank.
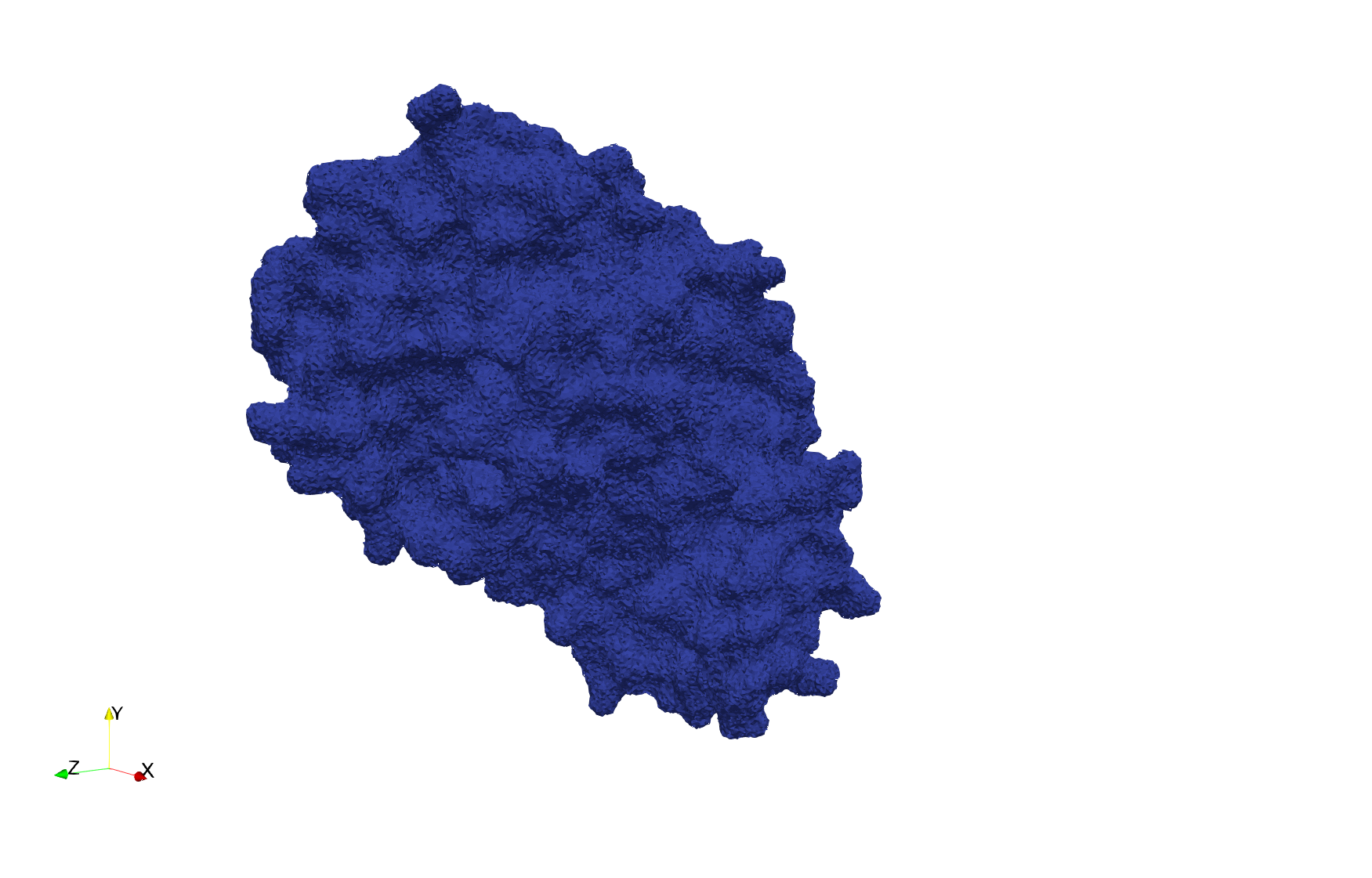
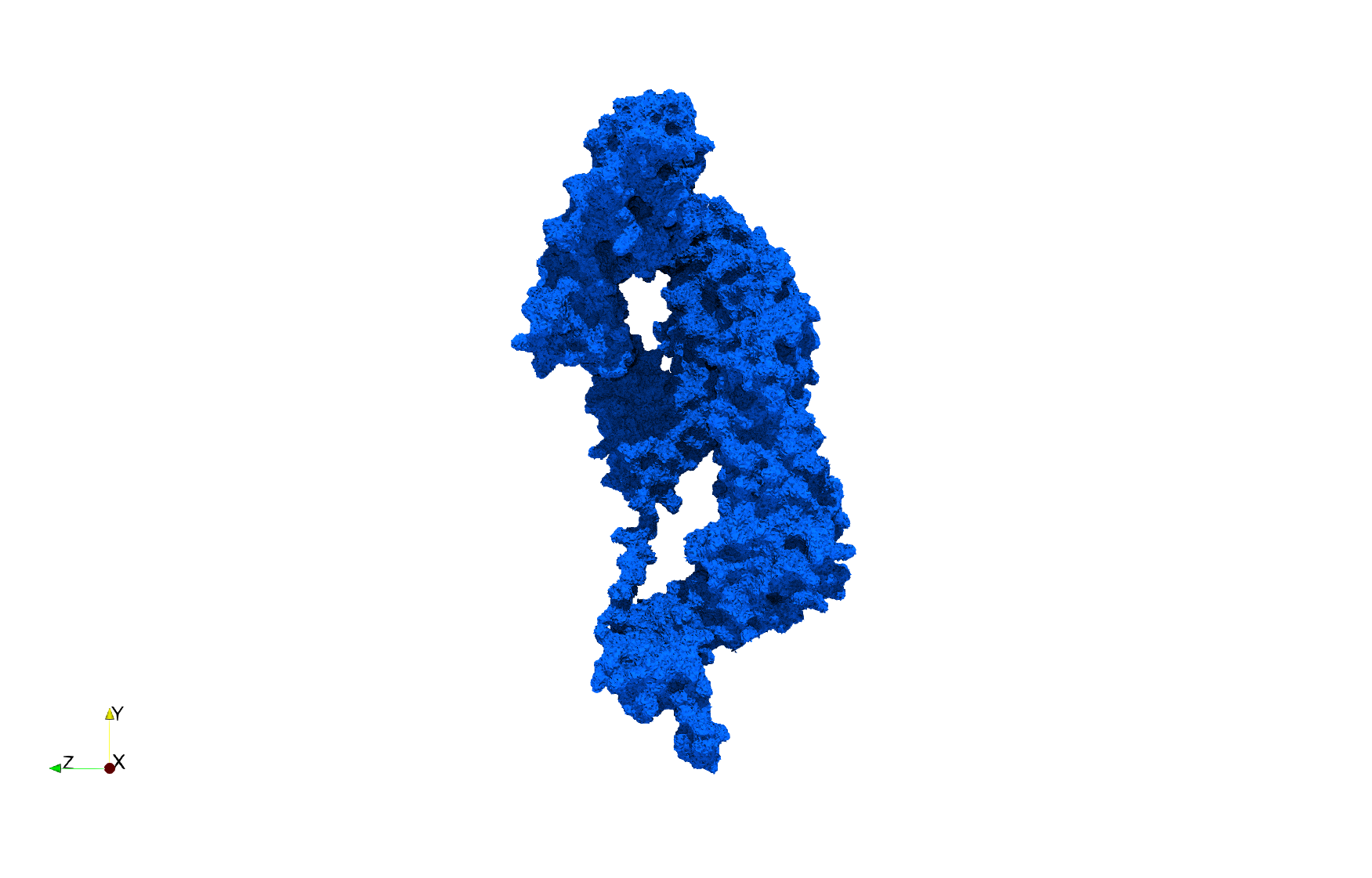
COVID-19-NSP-15-Endoribonuclease-6vww
- [Input Image]
- Input image : Dimensions (303x297x337) with spacing (0.3445713x0.3445713x0.3445713)
- Uniform with Delta = 0.5: 2,310,215 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-NSP-15-Endoribonuclease-6vww.nrrd --delta 0.5 --output ./COVID-19-NSP-15-Endoribonuclease-6vww,d=0.5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 NSP15 Endoribonuclease. Biological assembly data was retrieved from the molecule 6vww, Protein Data Bank.
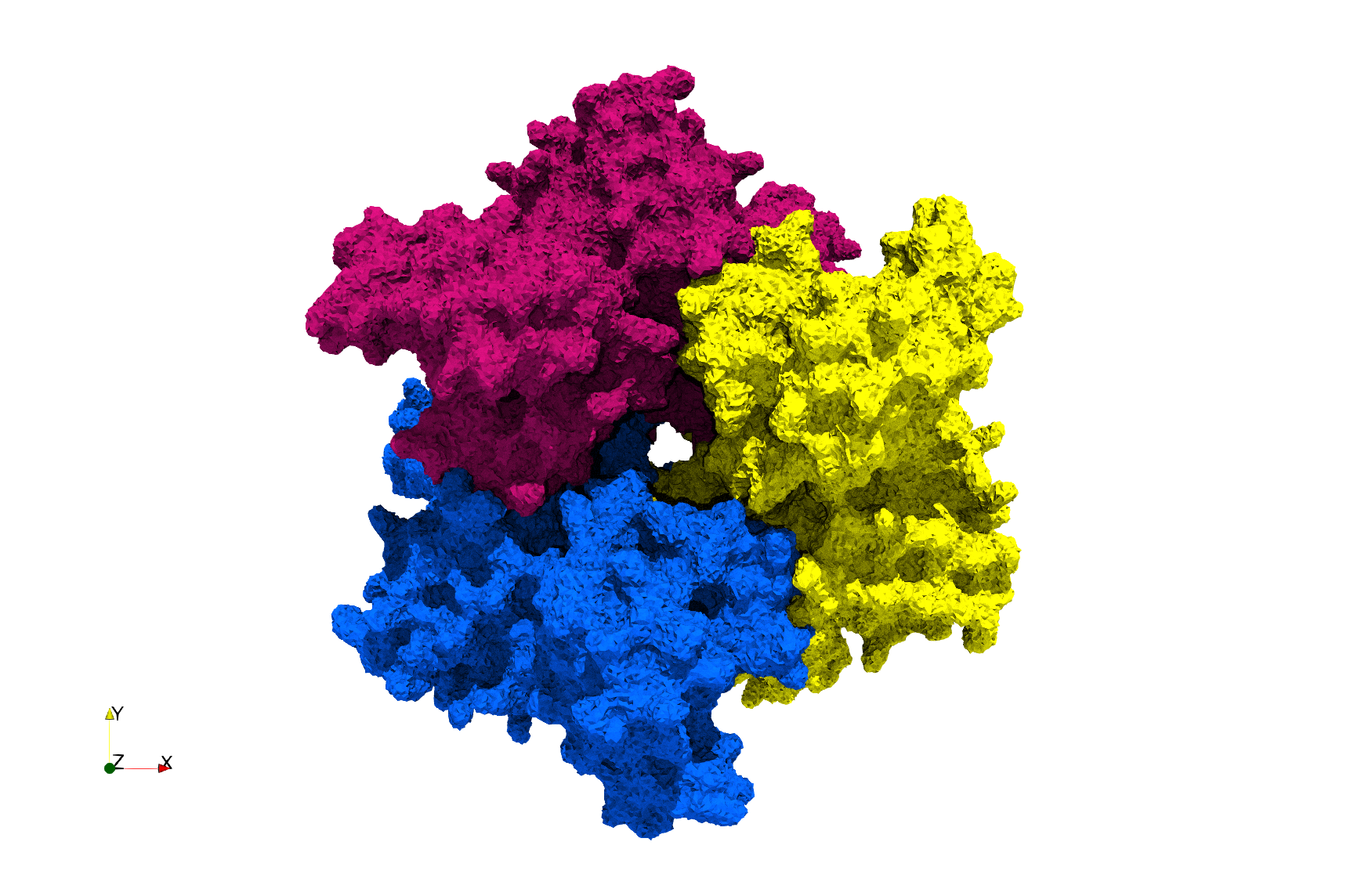
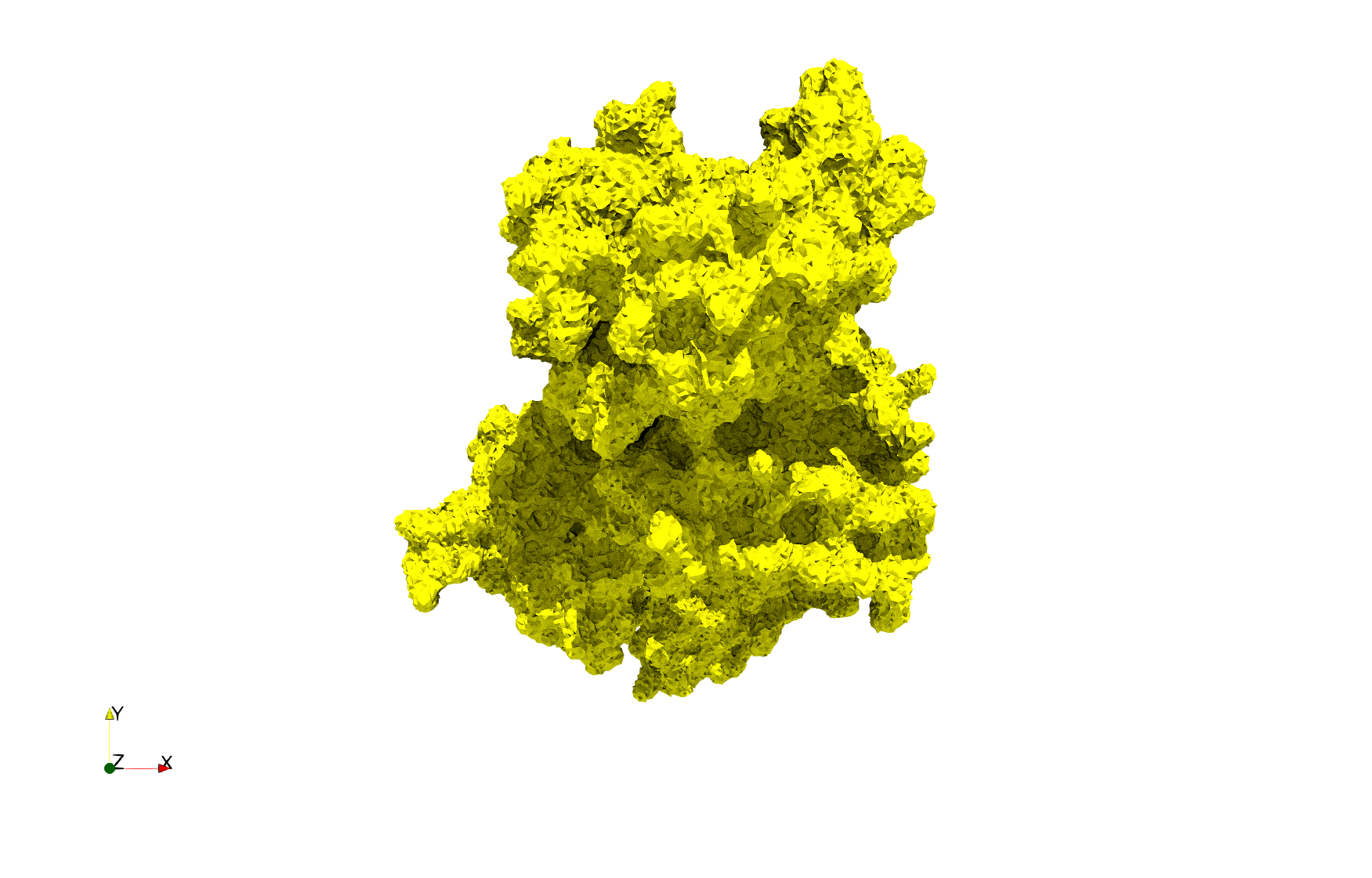
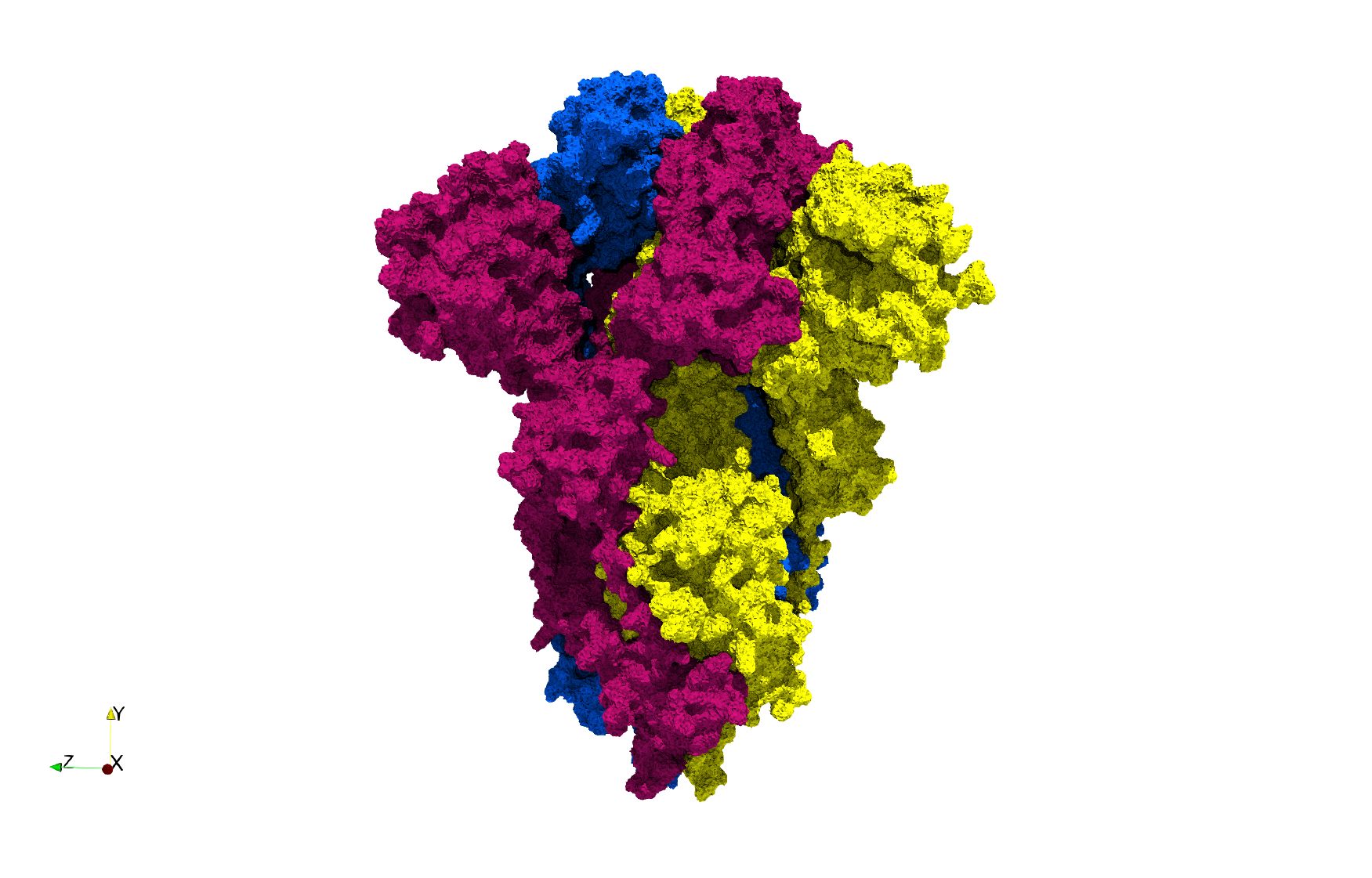
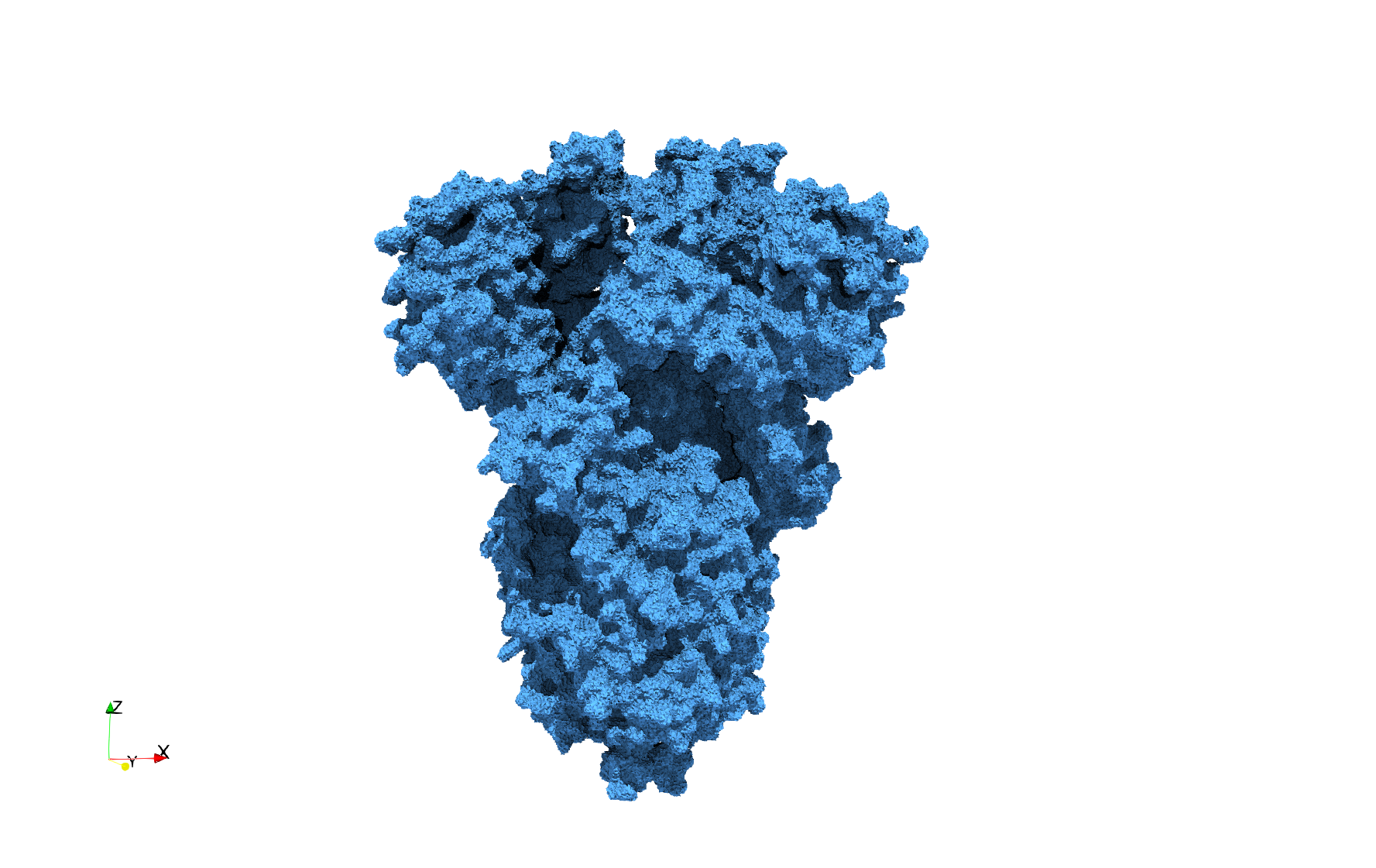
COVID-19-Spike-Glycoprotein-6vxx
- Input Image
- Input image : Dimensions (281x380x302) with spacing (0.427871x0.427871x0.427871)
- Uniform with Delta = 0.5: 3,260,055 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vxx.nrrd --delta 0.5 --output ./COVID-19-Spike-Glycoprotein-6vxx,d=5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 spike glycoprotein. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
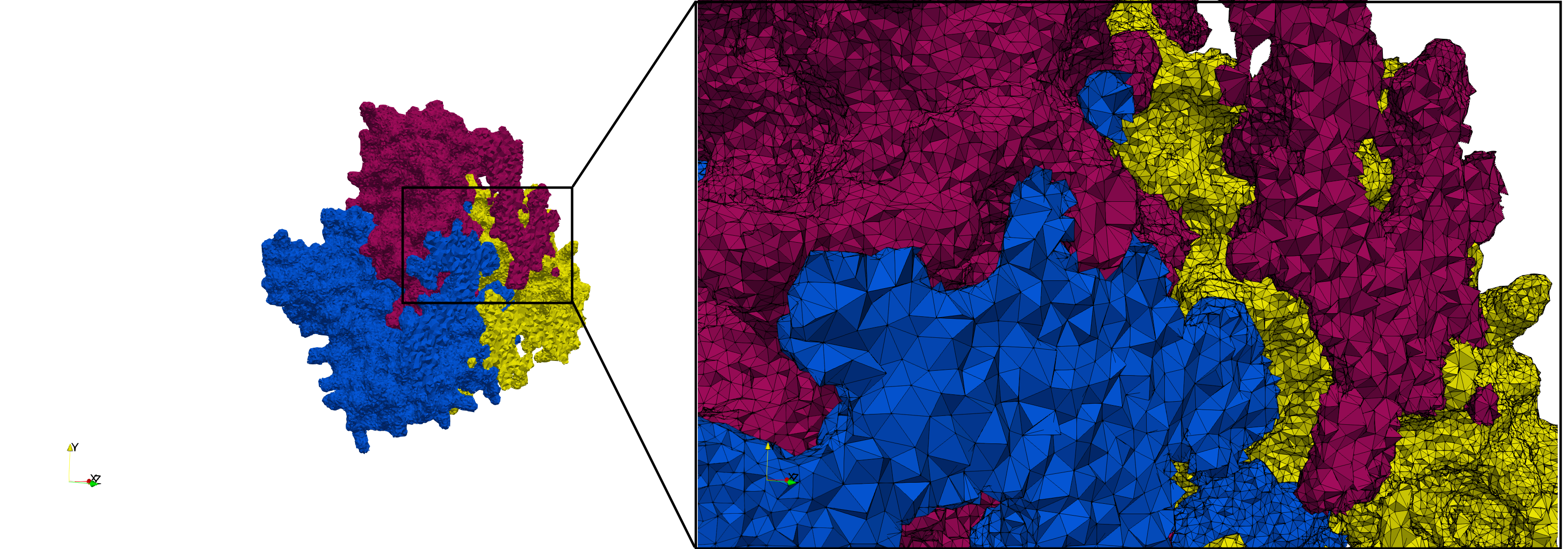
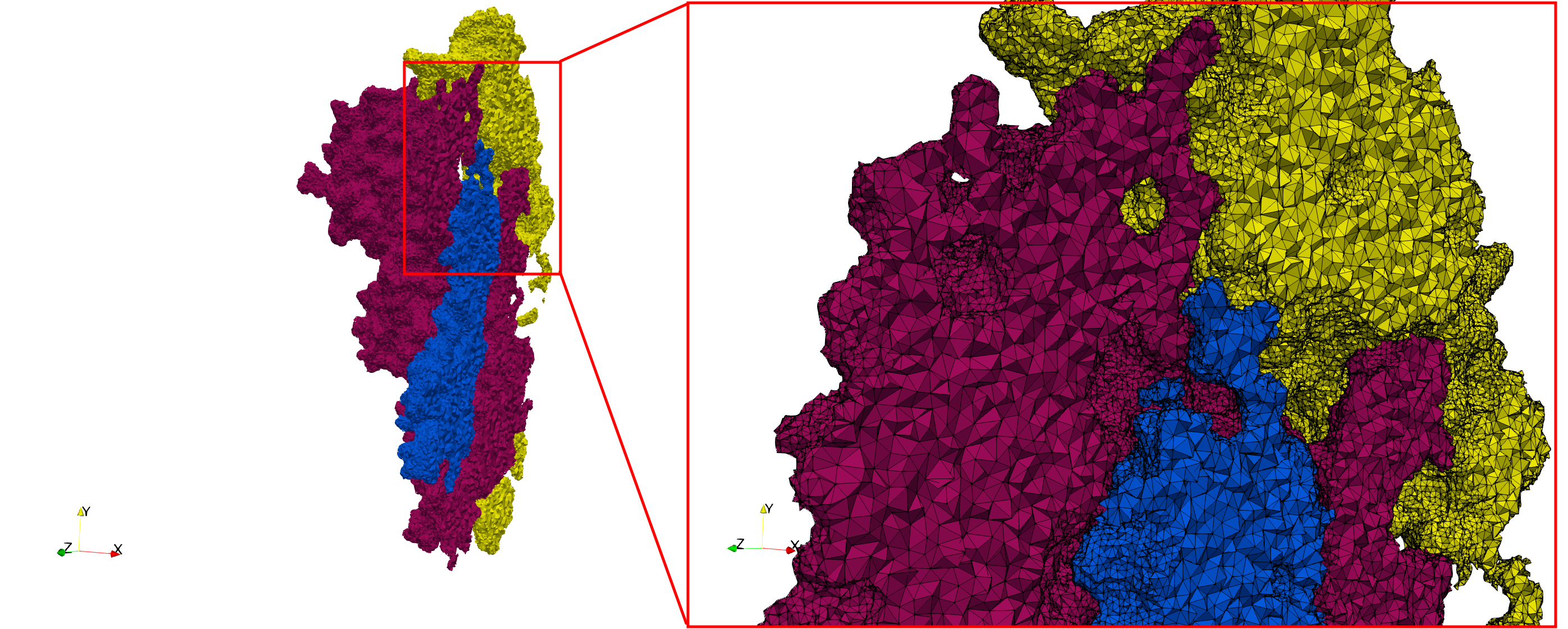
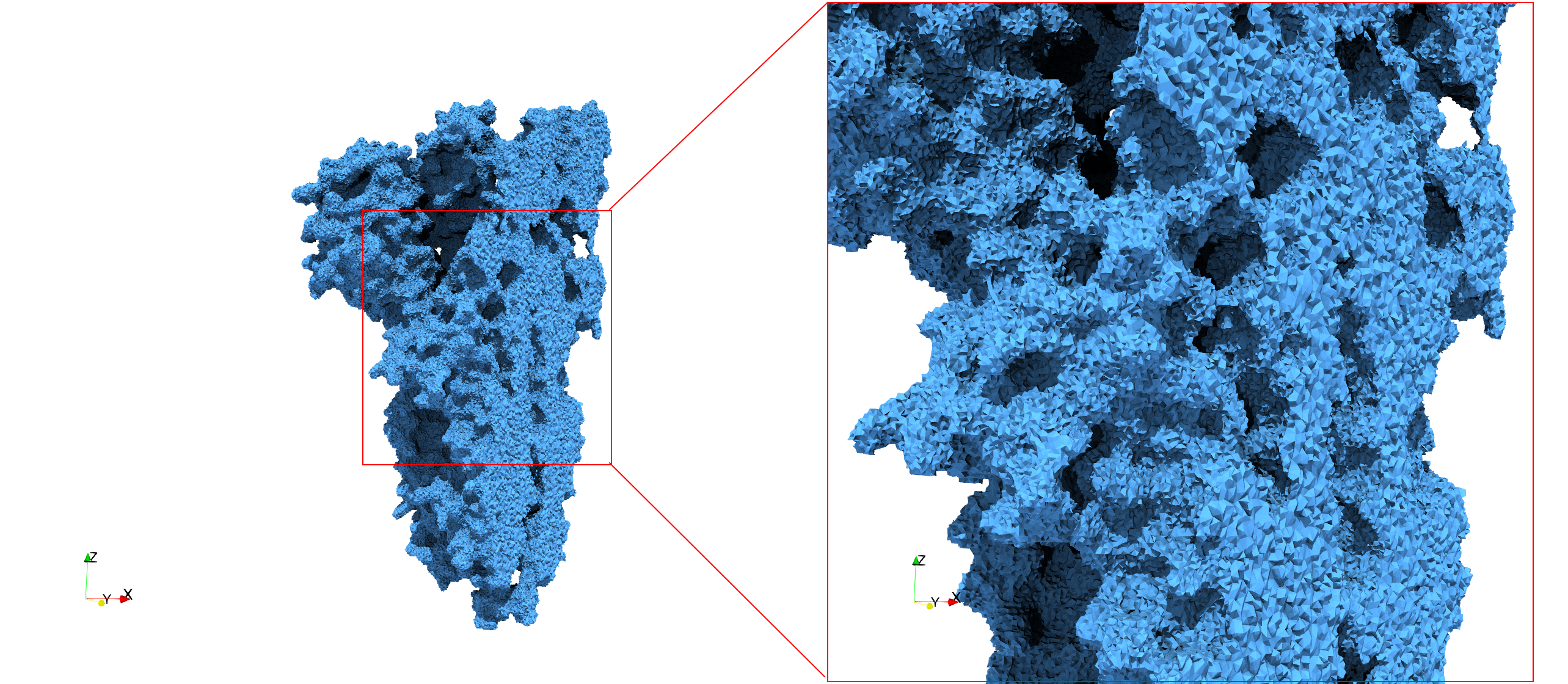
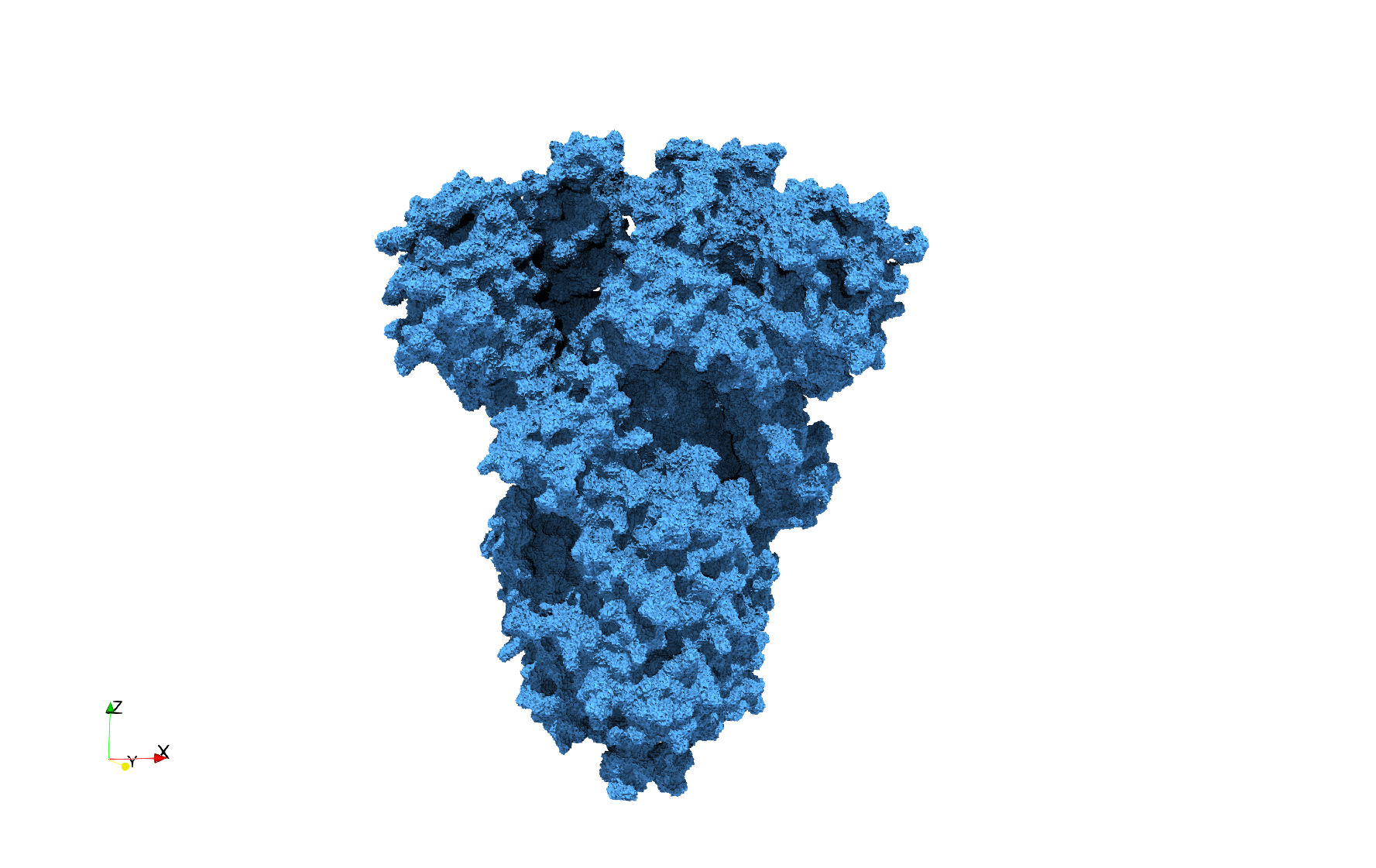
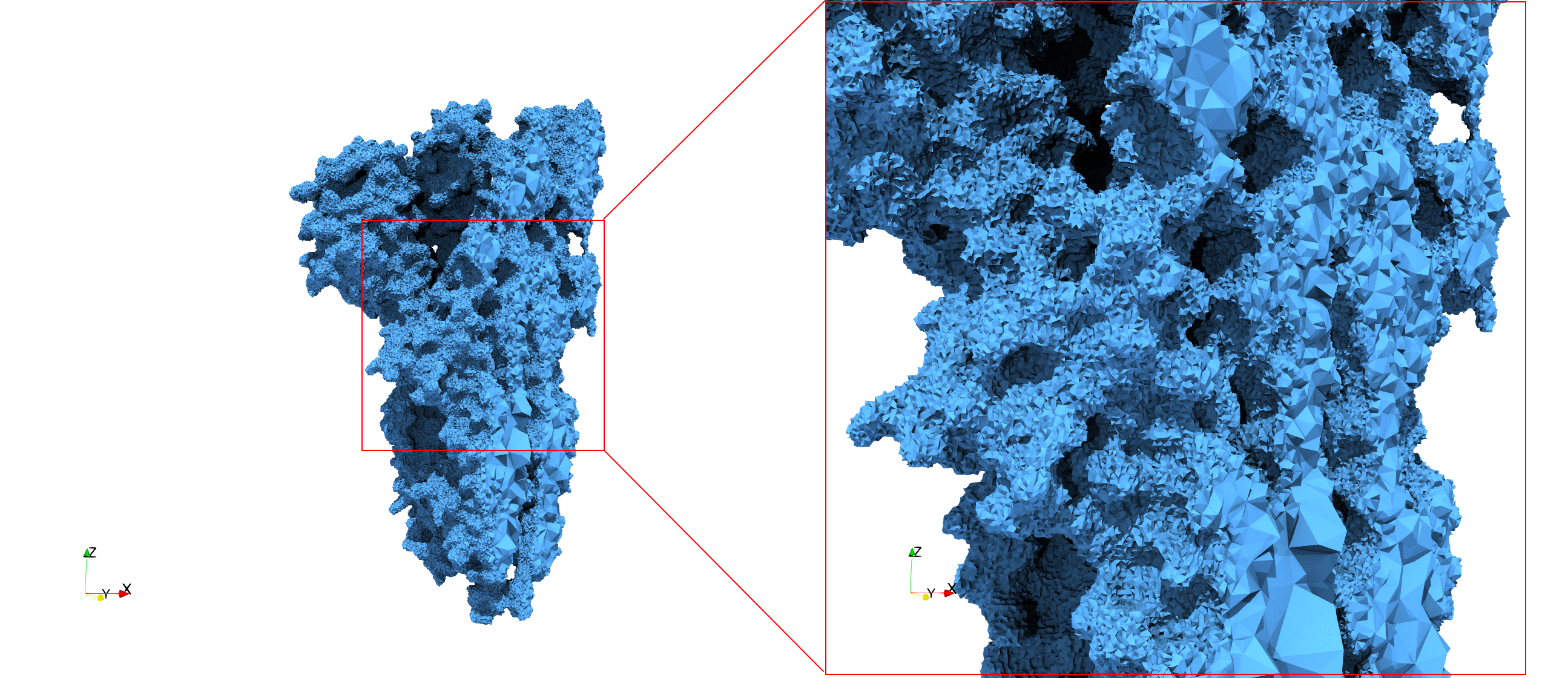
COVID-19-Spike-Glycoprotein-6vsb
- Input Image
- Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042)
- Uniform with Delta = 0.4: 5,731,833 tetrahedra
- Graded with Delta = 0.4: 3,794,223 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4.vtk
Graded with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --volume-grading --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.vtk
Uniform and graded tetrahedral meshes capable to help identify the internal voids of the COVID-19 spike glycoprotein. The middle and right meshes are slices. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
2D Example Meshes
The directory containing the 2D input data is located in the 2D folder of Bioninformatics_Data.
COVID-19-23311
- Input Image
- Input image : Dimensions (2,460x2,460) with spacing (1x1)
- Uniform with Min-Edge = 20: 47,028 triangles
- Adaptive with Min-Edge = 20: 34,080 tetrahedra
Commands to generate meshes:
Uniform with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --uniform --min-edge 20 --output ./COVID-19-23311,uniform,e=20.vtk
Adaptive with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --min-edge 20 --output ./COVID-19-23311,w=0.1,e=20.vtk
Meshes generated based on the image 23311 retrieved from the Centers for Disease Control and Prevention. For the uniform mesh, the edge-size corresponds to 20 pixels. The adaptive was created by controlling the size of the elements based on the difference in the intensity of the pixels.
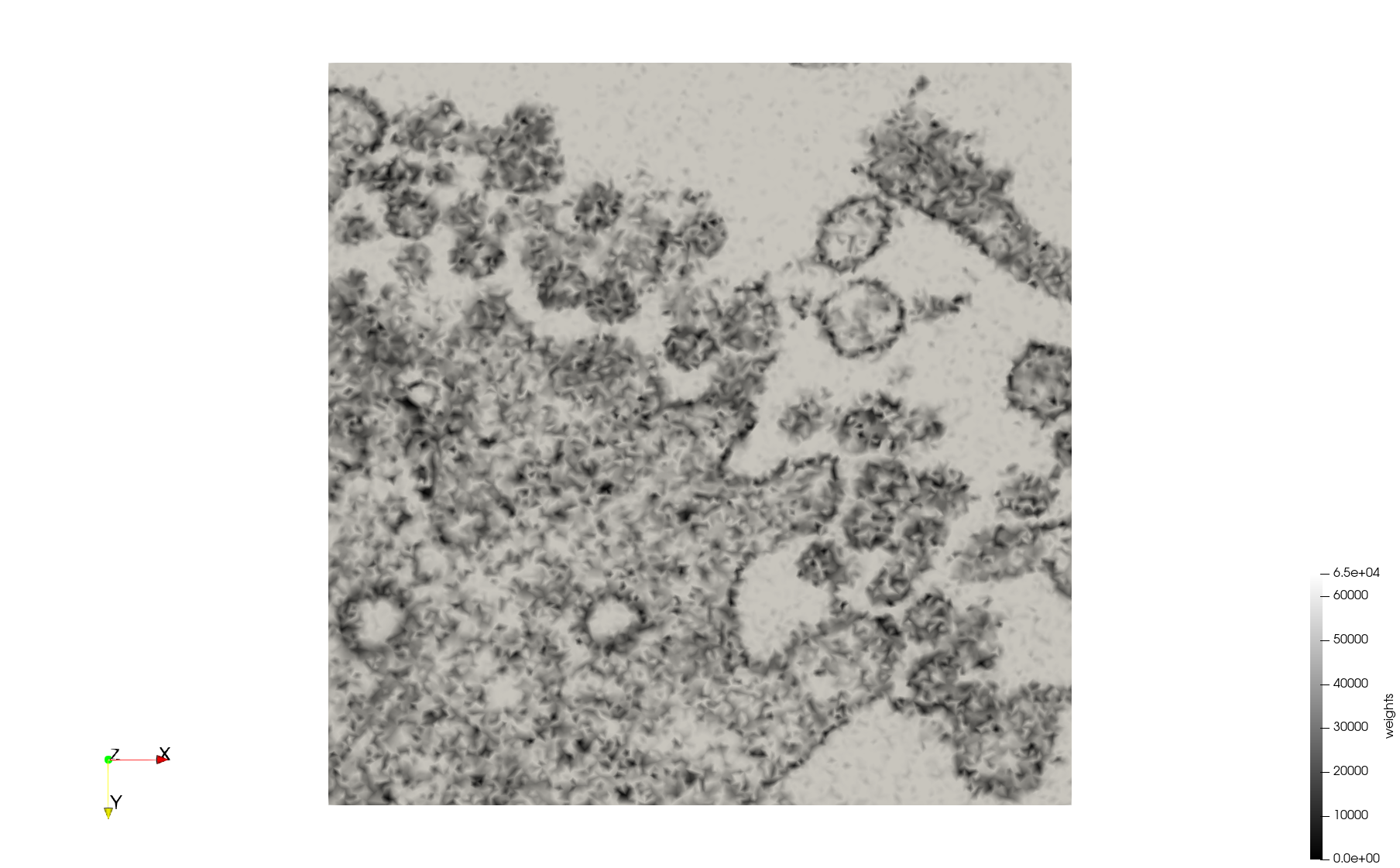
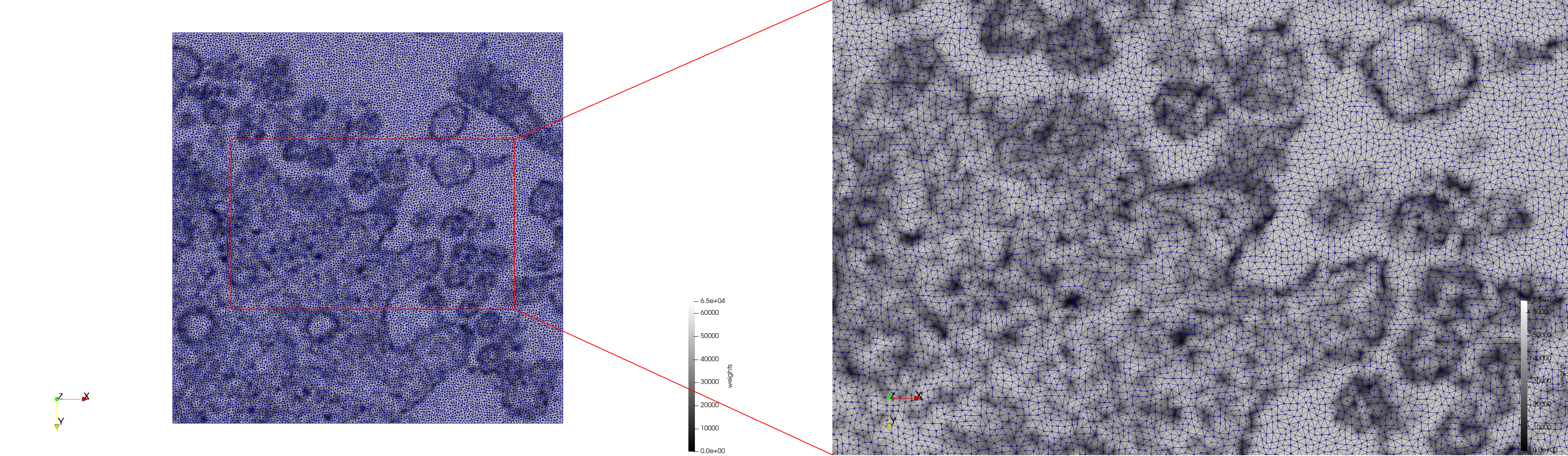
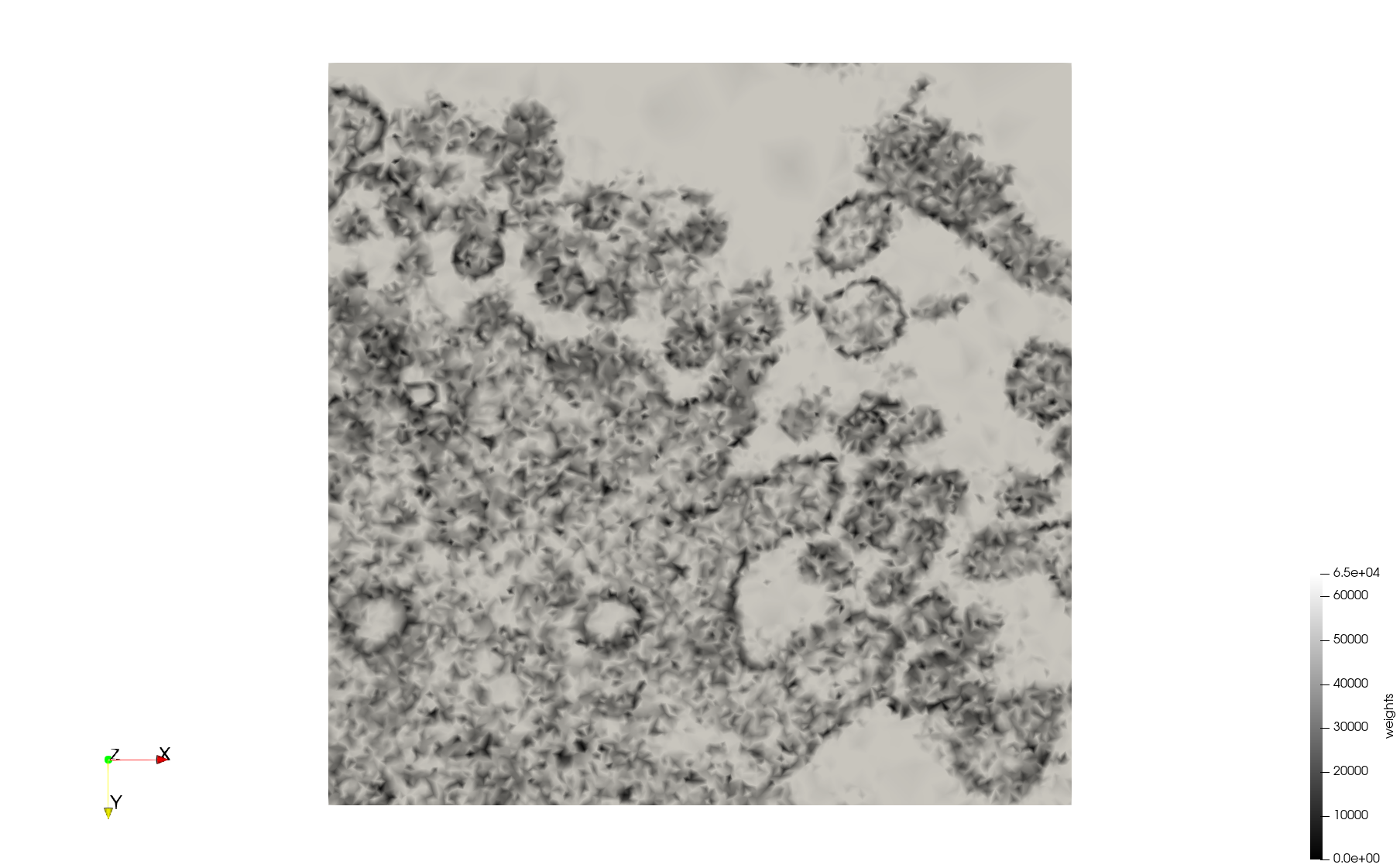
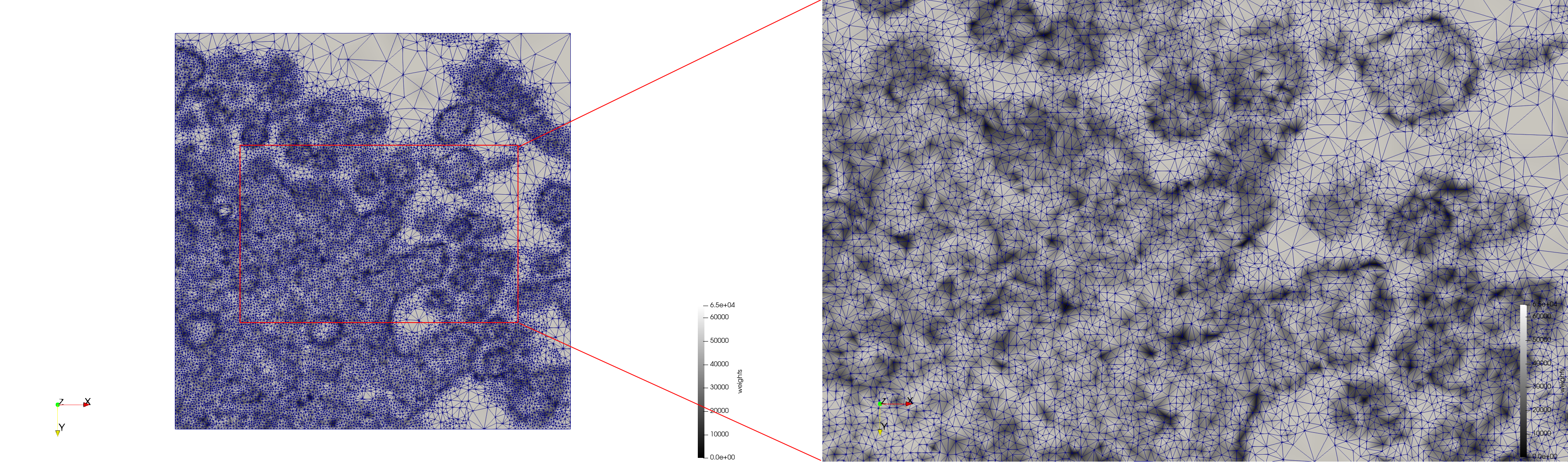
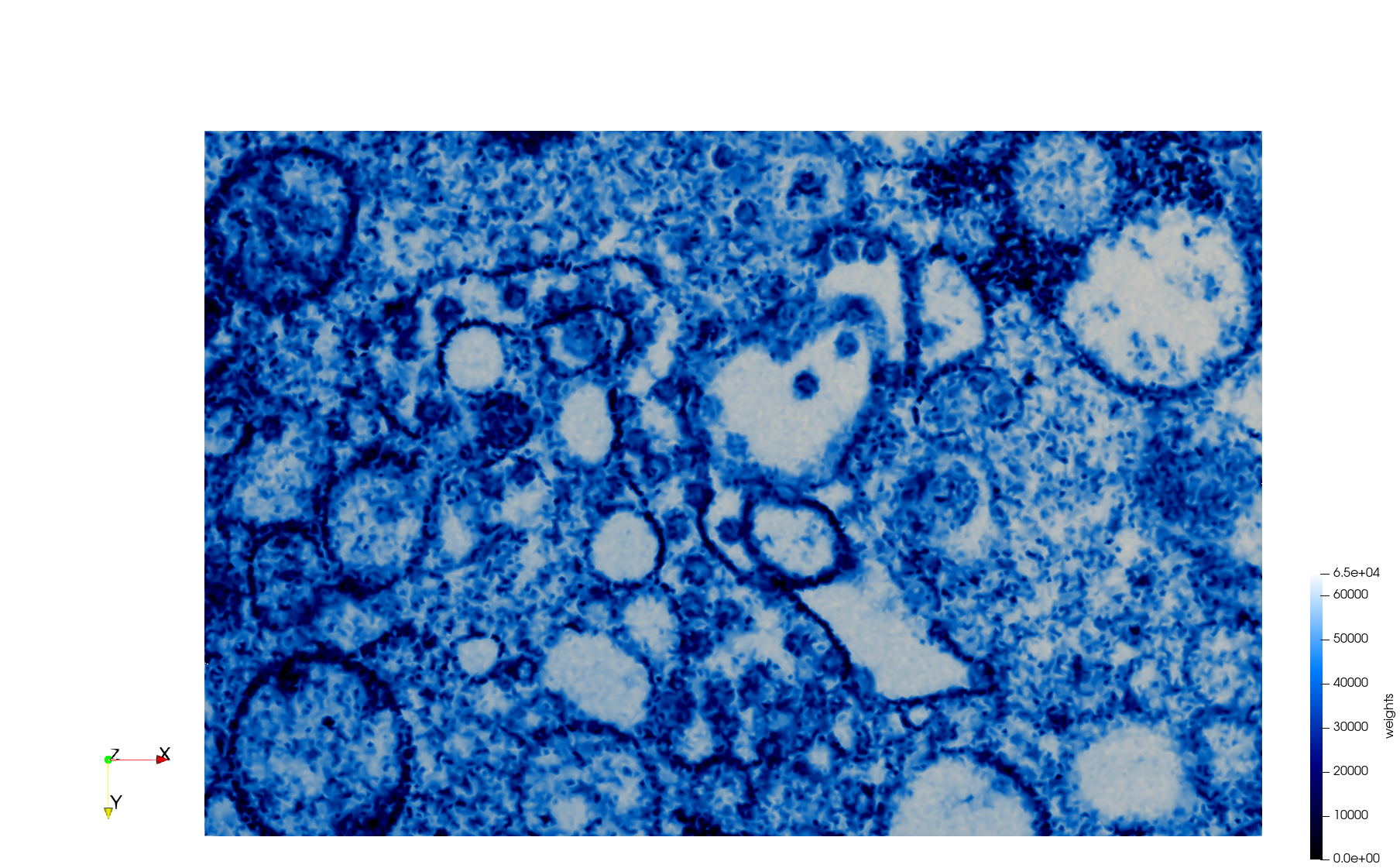
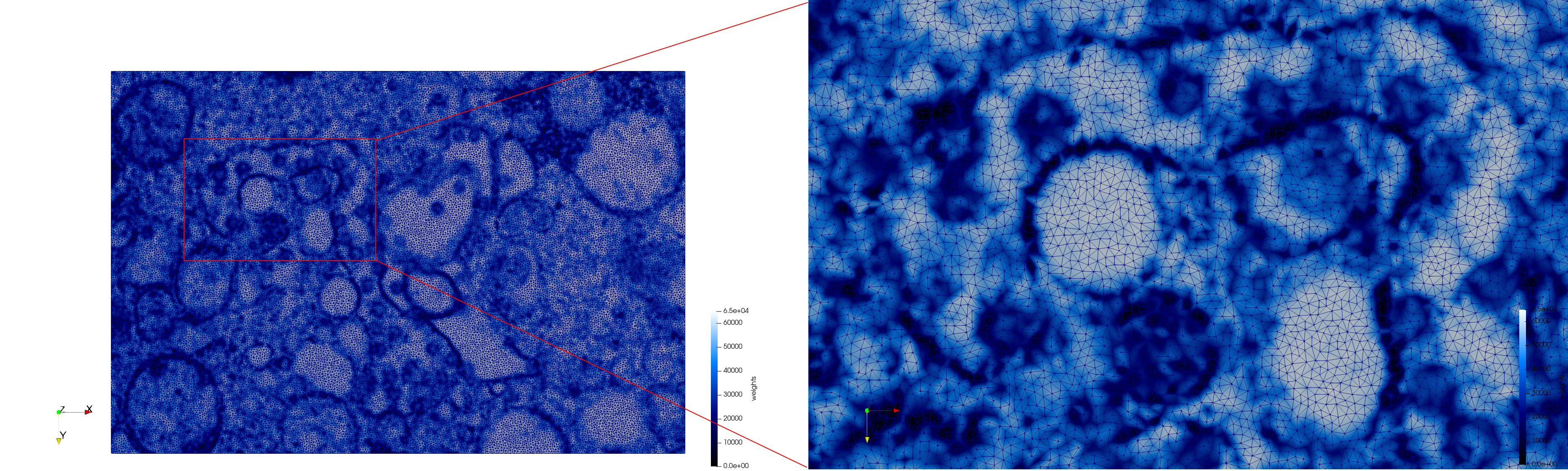
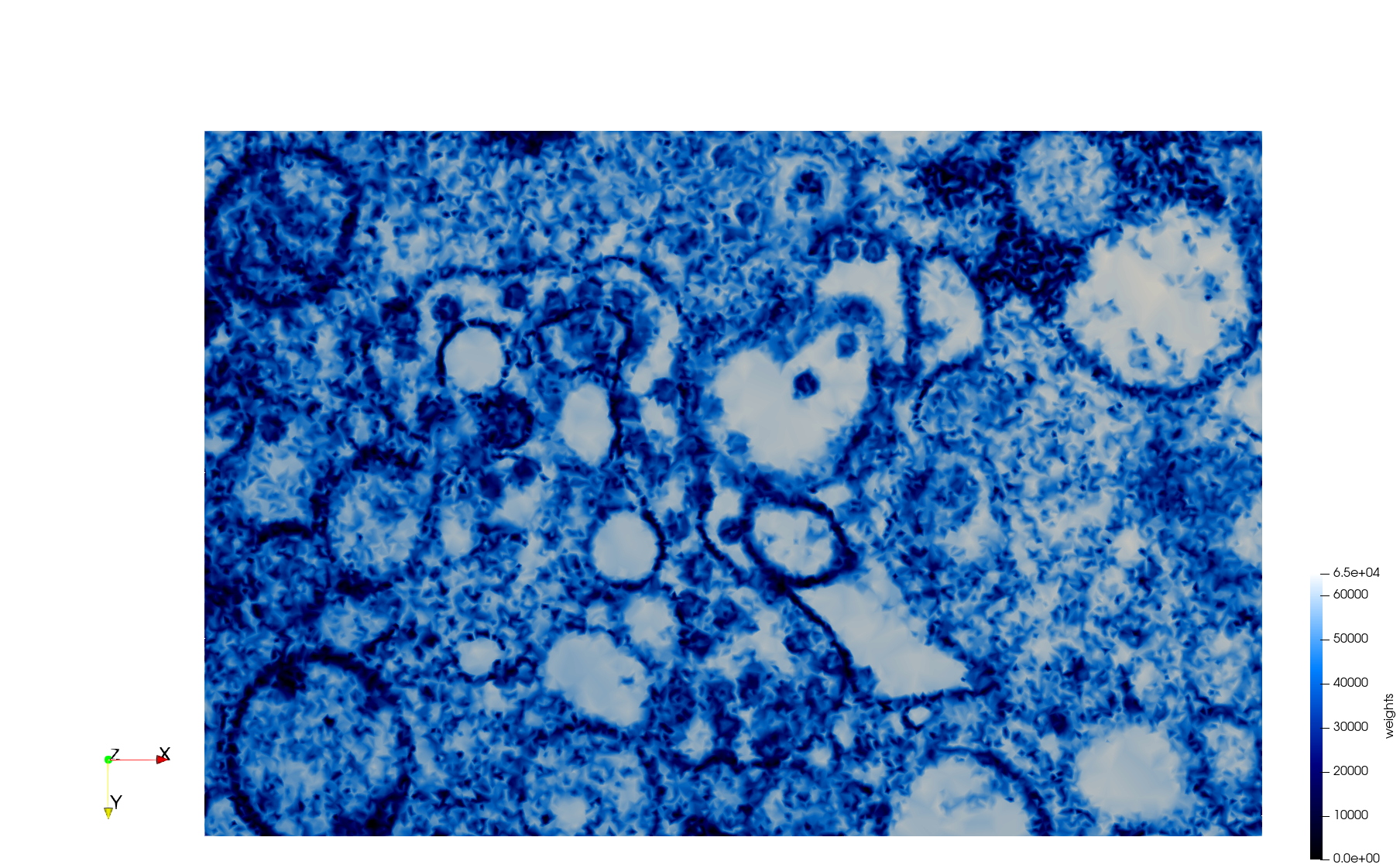
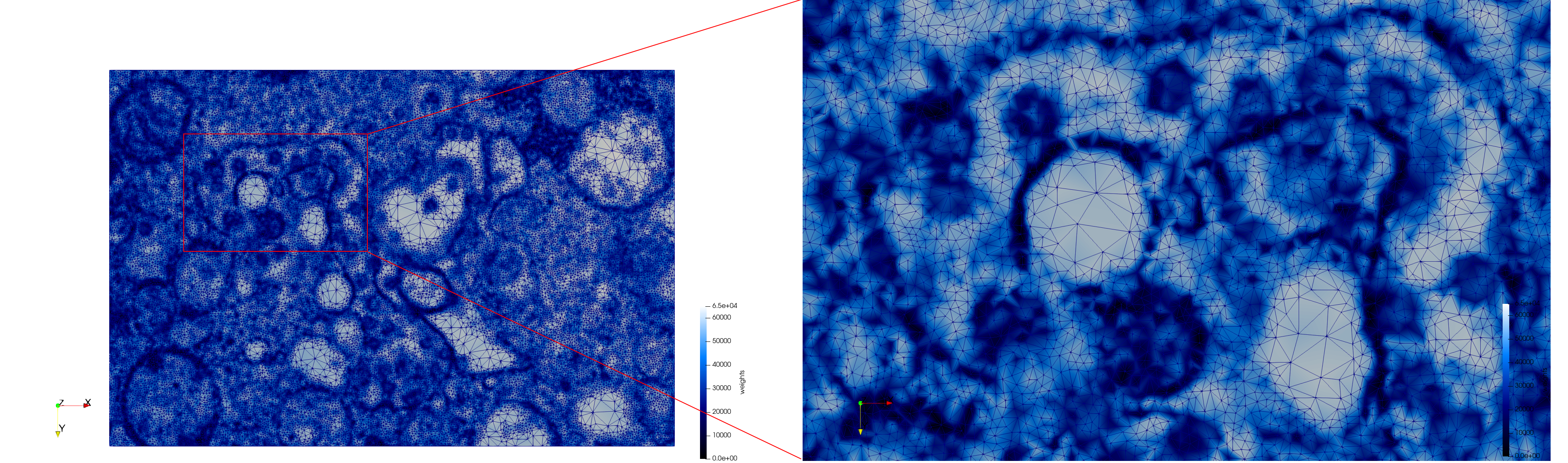
COVID-19-23354
- Input Image
- Input image : Dimensions (3,000x2,000) with spacing (1x1)
- Uniform with Min-Edge = 15: 82,981 triangles
- Adaptive with Min-Edge = 15: 67,920 triangles
Commands to generate meshes:
Uniform with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --uniform --min-edge 15 --output ./COVID-19-23354,uniform,e=15.vtk
Adaptive with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --min-edge 15 --output ./COVID-19-23354,w=0.1,e=15.vtk
Meshes generated based on the image 23354 retrieved from the Centers for Disease Control and Prevention. For the uniform mesh, the edge-size corresponds to 15 pixels. The adaptive was created by controlling the size of the elements based on the difference in the intensity of the pixels.