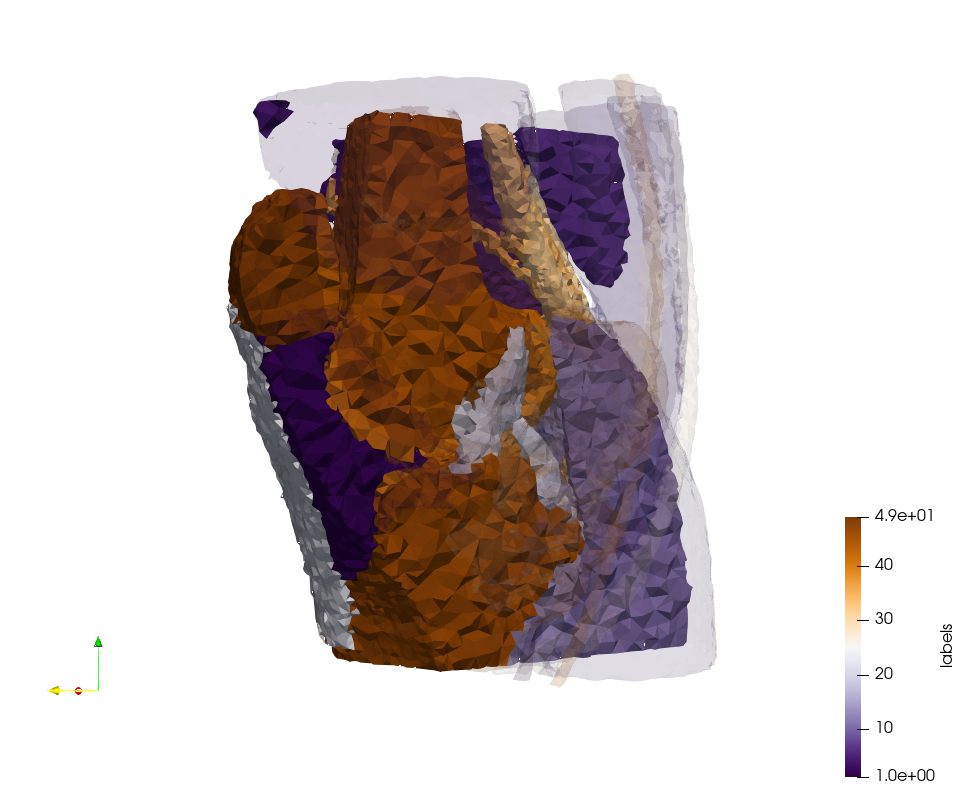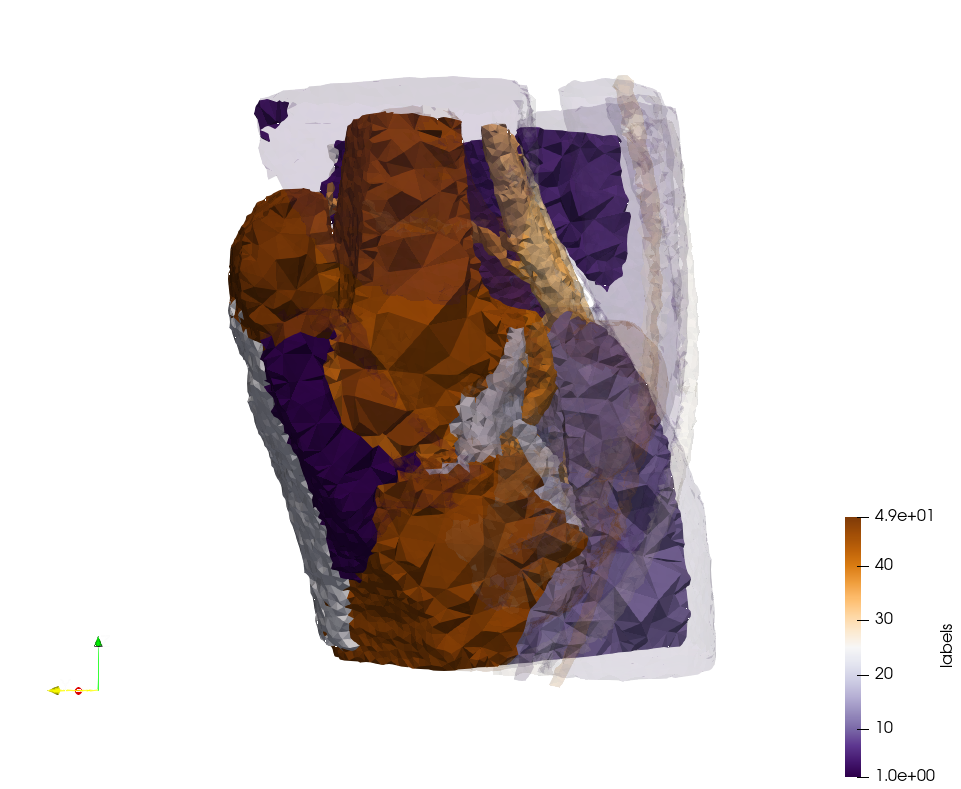Medical Imaging Example Meshes
From crtc.cs.odu.edu
Contents
Role of Delta
Table of produced amount elements for certain values of delta using the image: Brain-With-Tumor-Case17.
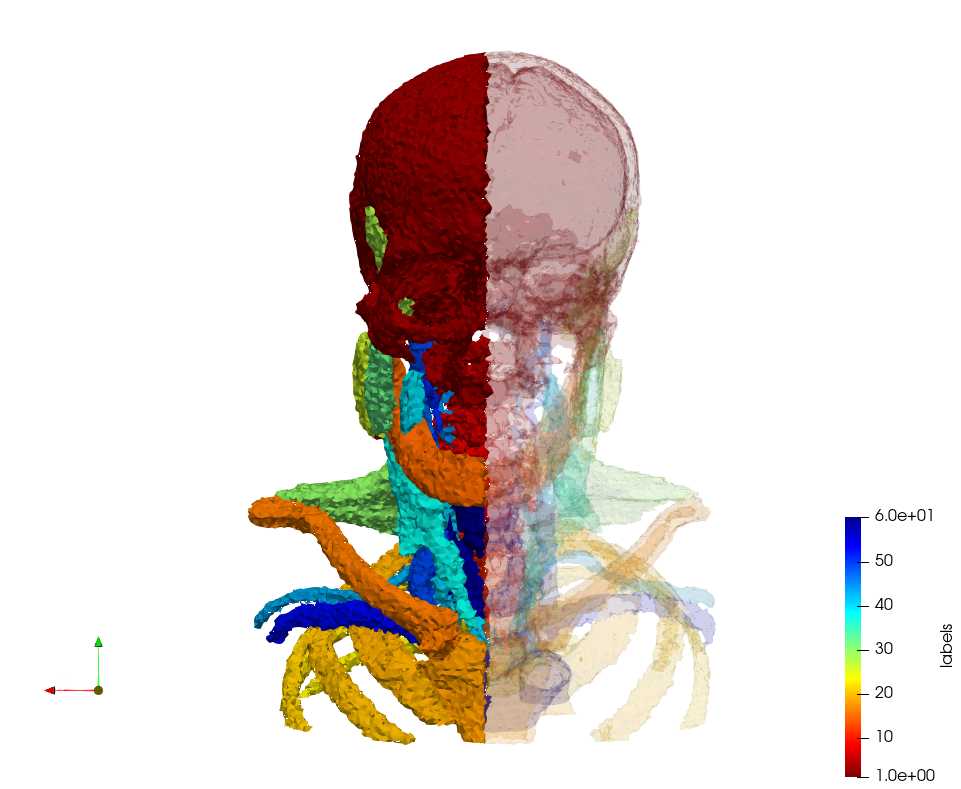
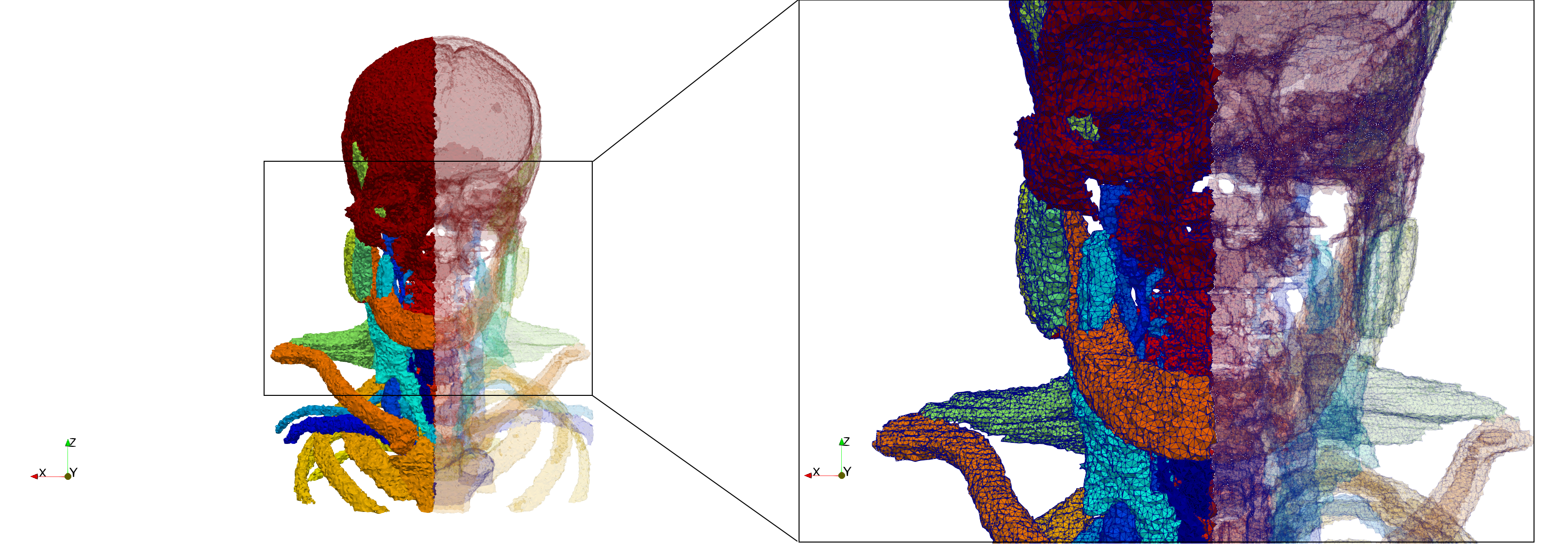
Head-Neck
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 2 --output ./Head-Neck,d=2.vtk
Generated mesh with delta = 1.5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
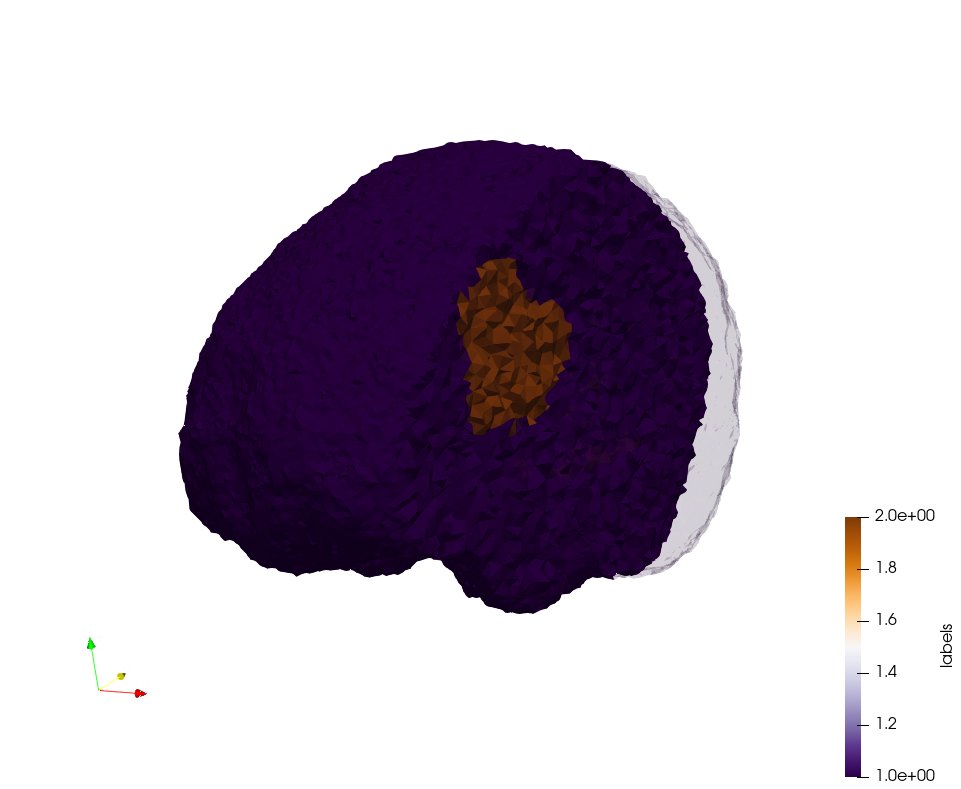
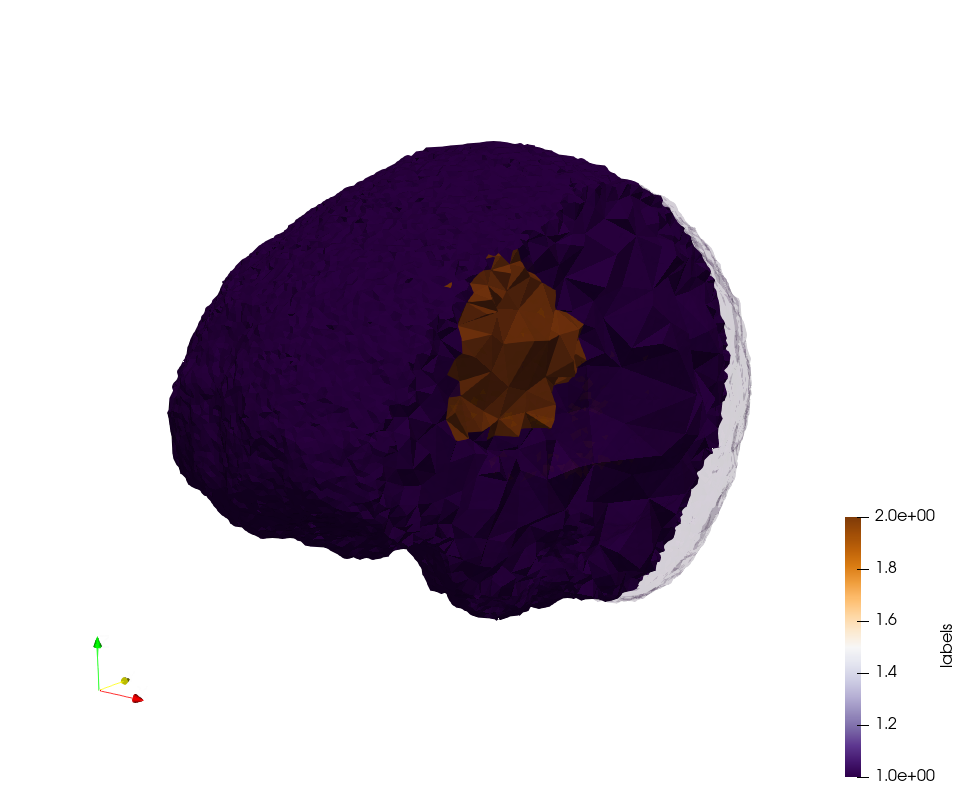
Brain-With-Tumor-Case17
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk
Knee-Char
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk
Big-Brain
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Big-Brain.mha --delta 2 --output ./Big-Brain,d=2.vtk
Ircad2-Removed-Tissues
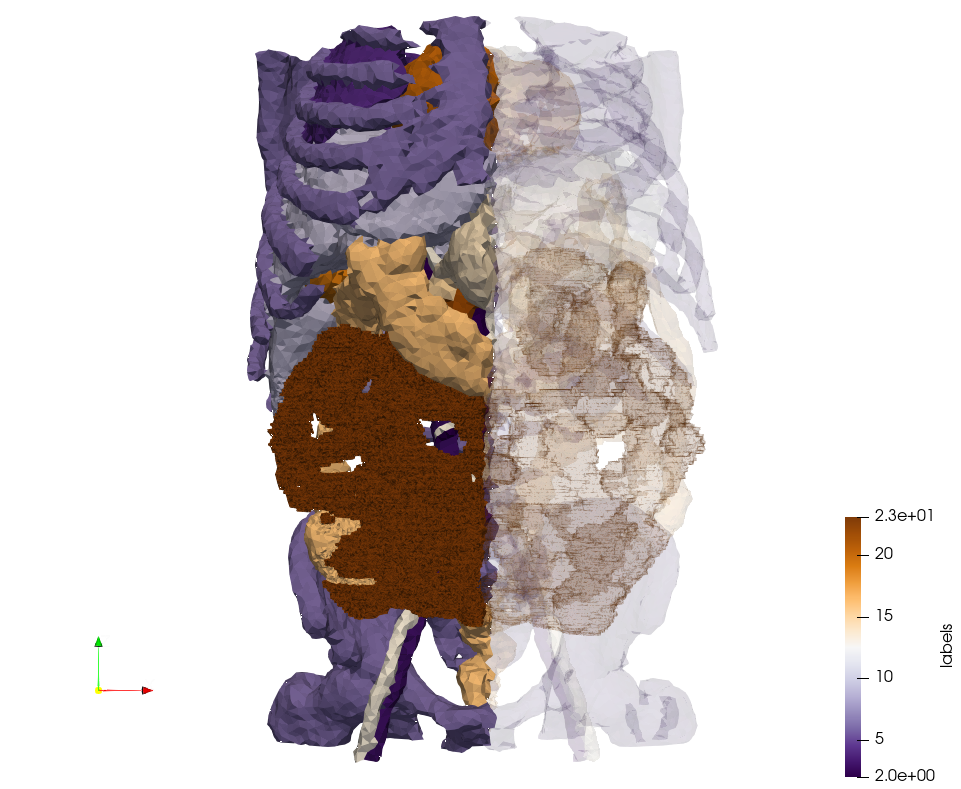
Generated mesh with delta = 5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk