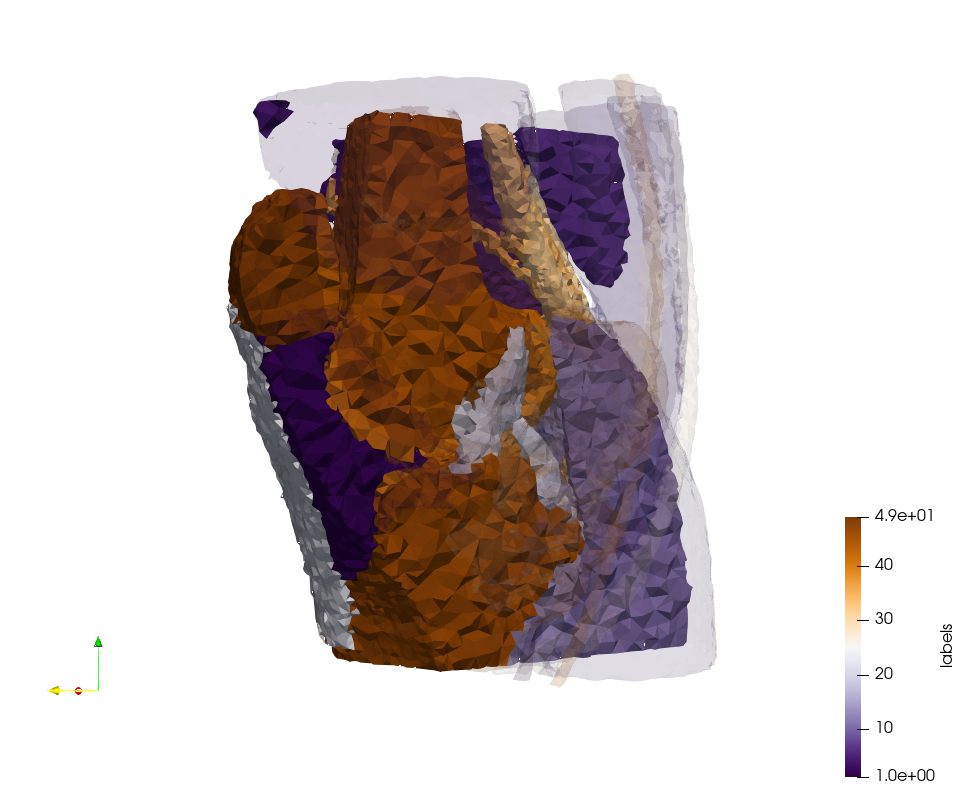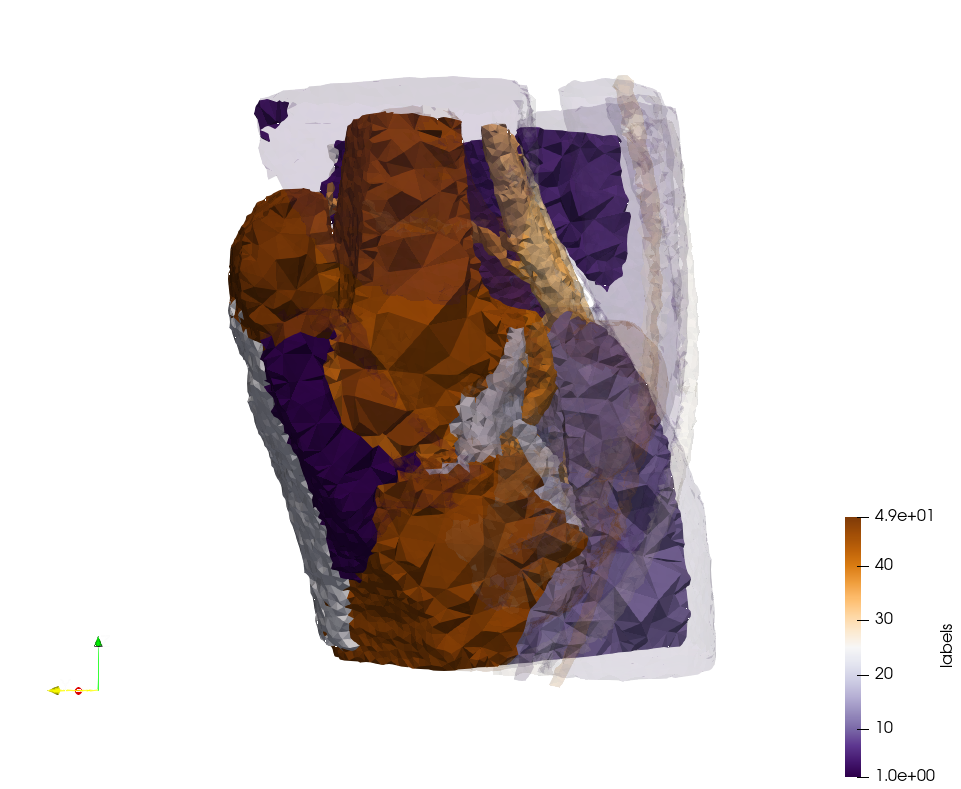Difference between revisions of "Medical Imaging Example Meshes"
From crtc.cs.odu.edu
Spyridon97 (talk | contribs) (→Role of Delta) |
Spyridon97 (talk | contribs) (→Role of Delta) |
||
| Line 1: | Line 1: | ||
=Role of Delta= | =Role of Delta= | ||
| + | Input Image: Knee-Char.mha | ||
The size of the input image is: (512x512x119) | The size of the input image is: (512x512x119) | ||
Spacing of the input image: (0.27734,0.27734,1) | Spacing of the input image: (0.27734,0.27734,1) | ||
| Line 7: | Line 8: | ||
Max delta value that possibly will not cause problems is: 119 / 5 = 23.8 | Max delta value that possibly will not cause problems is: 119 / 5 = 23.8 | ||
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
! Delta !! # Vertices !! # Tetrahedra | ! Delta !! # Vertices !! # Tetrahedra | ||
Revision as of 22:29, 19 February 2020
Contents
Role of Delta
Input Image: Knee-Char.mha The size of the input image is: (512x512x119) Spacing of the input image: (0.27734,0.27734,1)
MinimumPhysicalSize = 119 * 1 = 119. Max delta value that possibly will not cause problems is: 119 / 5 = 23.8
| Delta | # Vertices | # Tetrahedra |
|---|---|---|
| 23.8 | 70 | 153 |
| 20 | 87 | 222 |
| 15 | 219 | 668 |
| 10 | 551 | 1580 |
| 5 | 2858 | 10073 |
| 4 | 4544 | 16569 |
| 3 | 9279 | 35131 |
| 2 | 24631 | 98822 |
| 1.5 | 50371 | 209659 |
| 1 | 139515 | 613799 |
| 0.5 | 830423 | 3999734 |
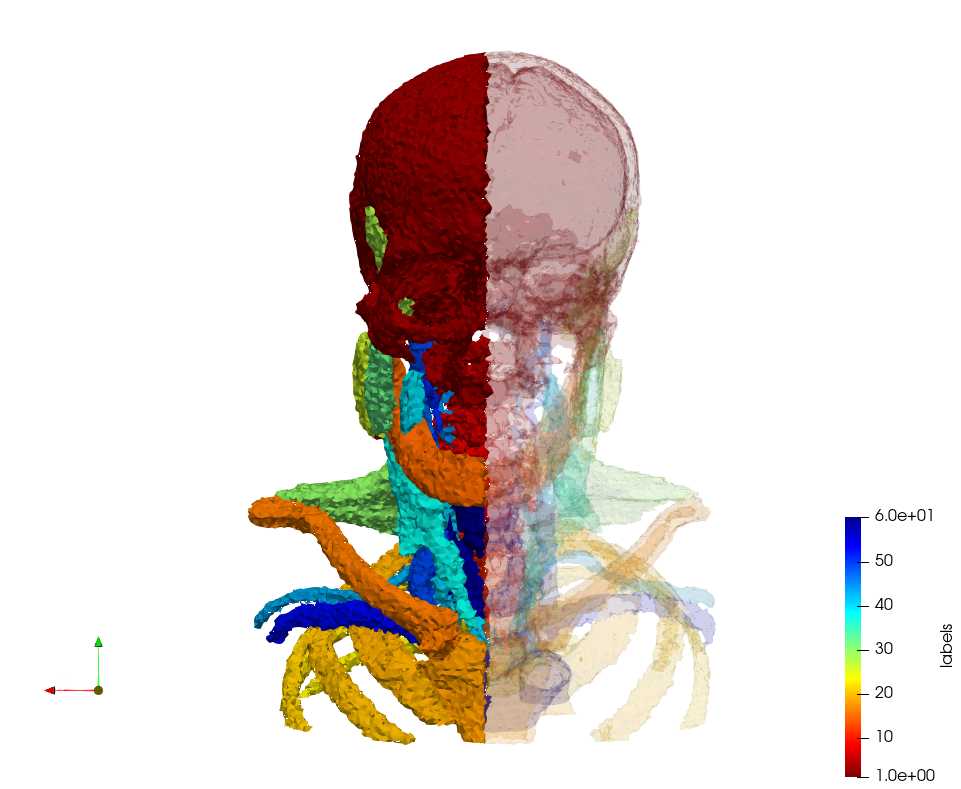
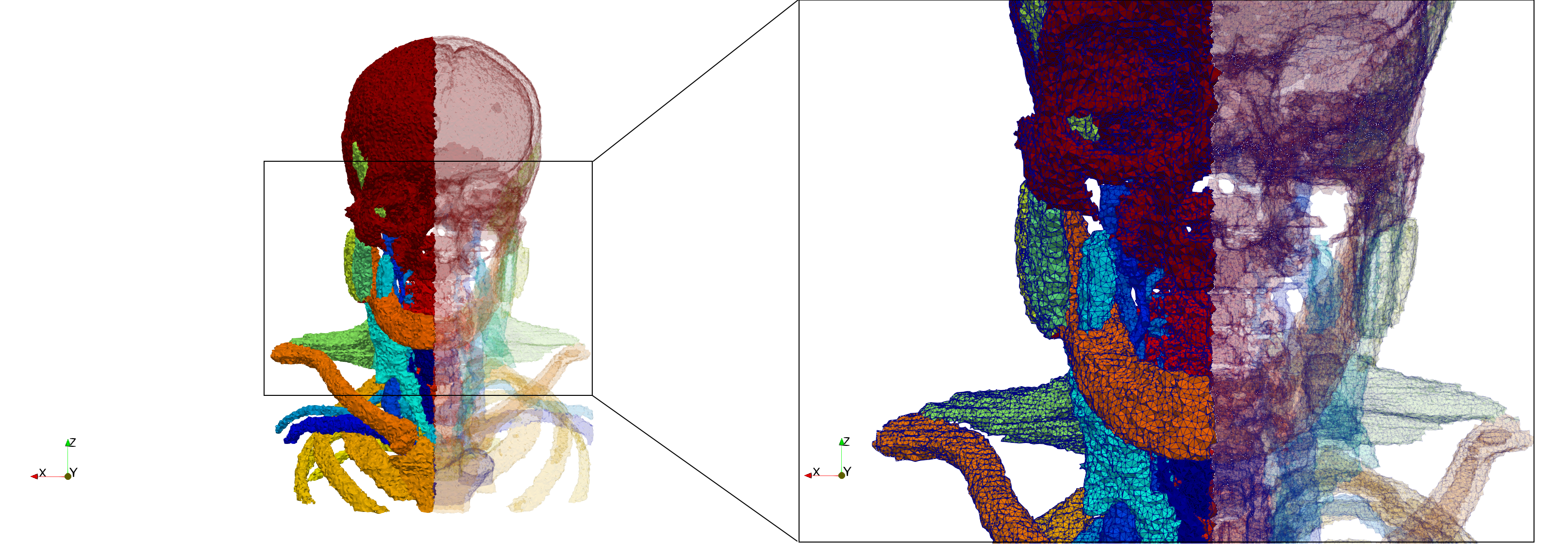
Head-Neck
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 2 --output ./Head-Neck,d=2.vtk
Generated mesh with delta = 1.5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
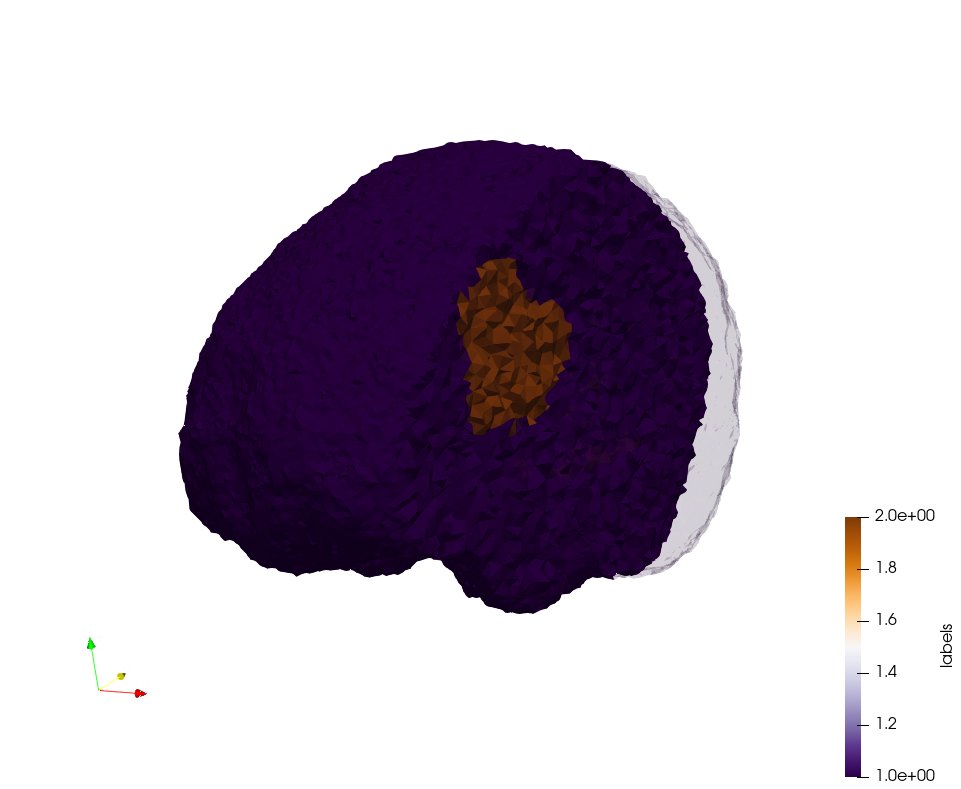
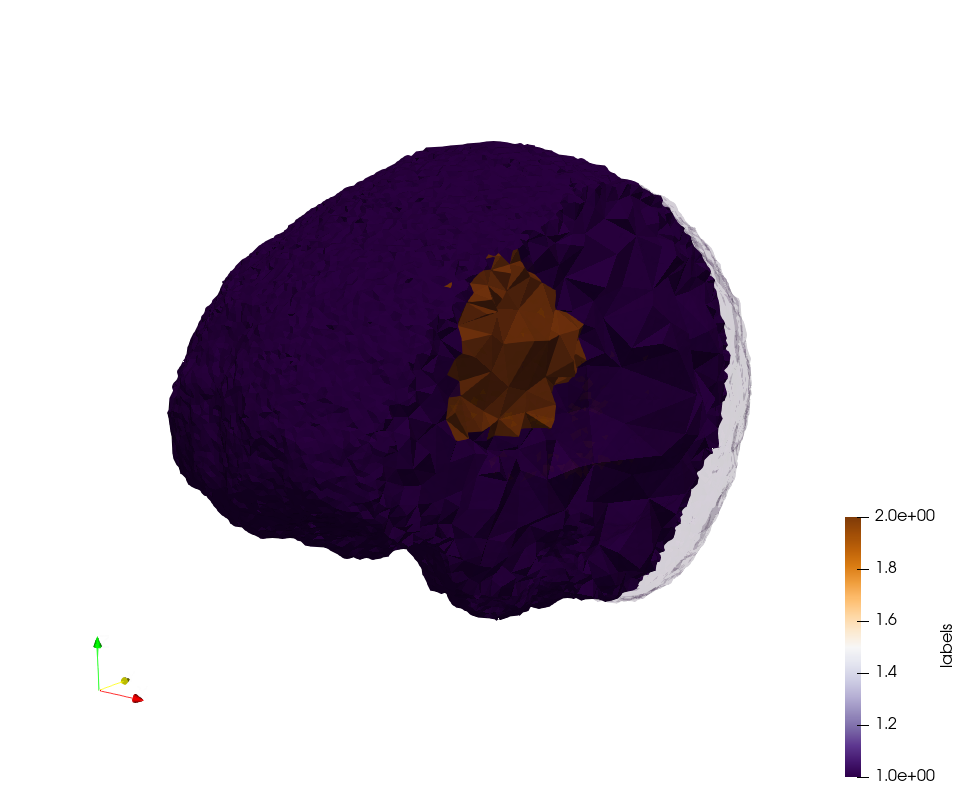
Brain-With-Tumor-Case17
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk
Knee-Char
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk
Big-Brain
Generated mesh with delta = 2, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Big-Brain.mha --delta 2 --output ./Big-Brain,d=2.vtk
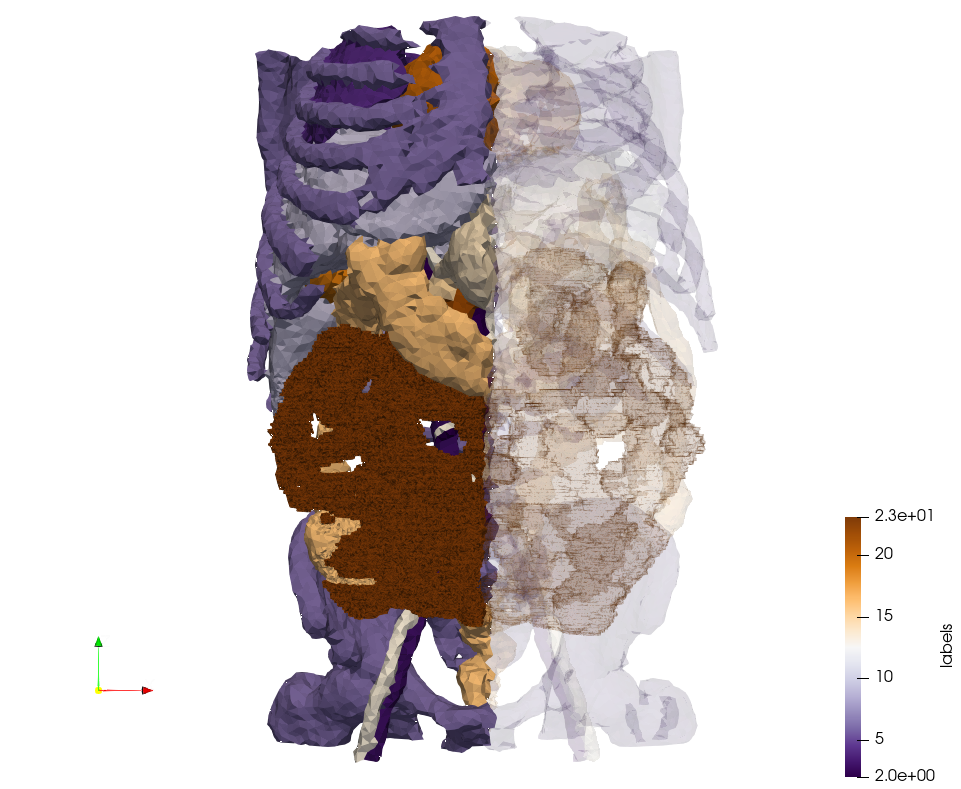
Ircad2-Removed-Tissues
Generated mesh with delta = 5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./NRL_SHARE/Ircad2-Removed-Tissues.nrrd --delta 5 --output ./Ircad2-Removed-Tissues,d=5.vtk