Difference between revisions of "Bioinformatics Example Meshes"
Spyridon97 (talk | contribs) (Created page with "__TOC__ =3D Example Meshes= The directory containing the 3D input data is located in the 3D folder of [https://odu.box.com/s/olefferrnksu2nmerbfvbvsz2u4abbdw Bioninformatics_...") |
Spyridon97 (talk | contribs) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
=3D Example Meshes= | =3D Example Meshes= | ||
| − | The directory containing the 3D input data is located in the 3D folder of [https://odu.box.com/s/ | + | The directory containing the 3D input data is located in the 3D folder of [https://odu.box.com/s/9scj74ix4eulpvgc93sbwte1uvlde8b6 Bioninformatics_Data]. |
==COVID-19-Main-Protease-6y2e== | ==COVID-19-Main-Protease-6y2e== | ||
| Line 104: | Line 104: | ||
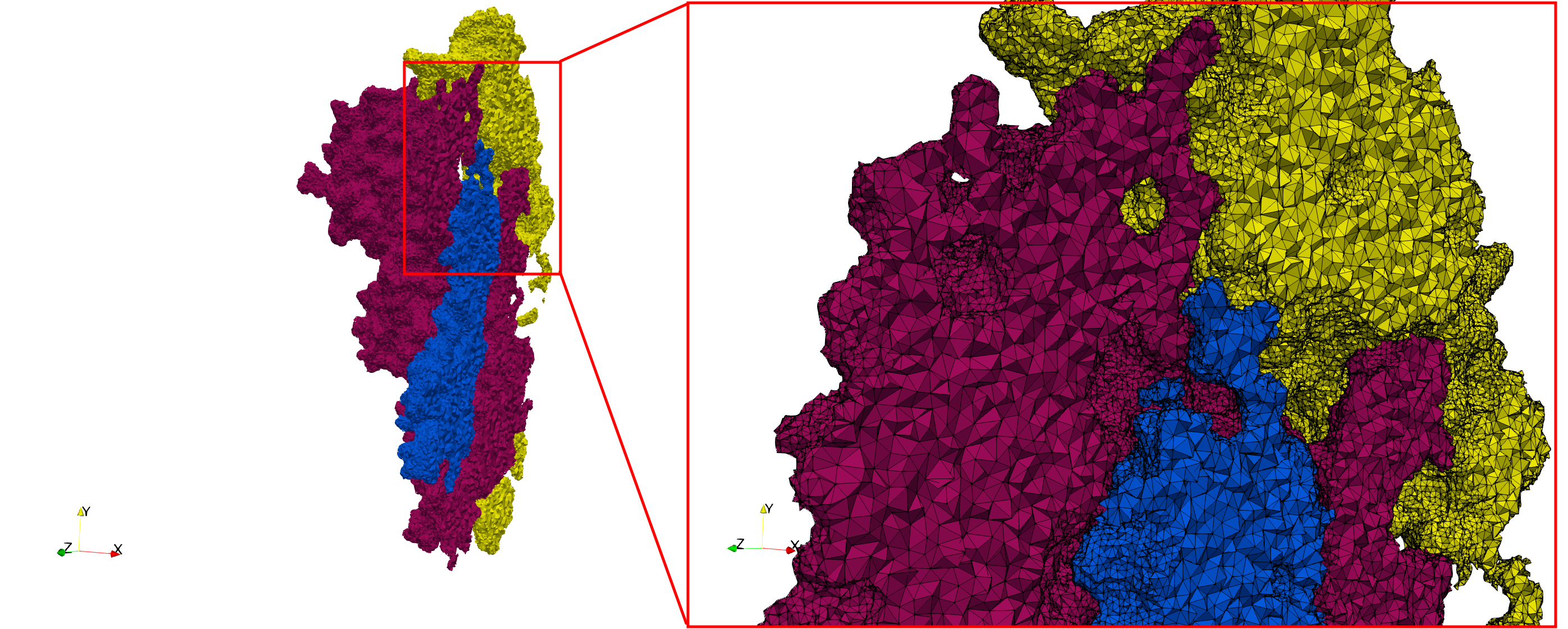
Uniform and graded tetrahedral meshes capable to help identify the internal voids of the <b>COVID-19 spike glycoprotein. </b> The middle and right meshes are slices. Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vxx 6vxx], [https://www.rcsb.org/ Protein Data Bank]. | Uniform and graded tetrahedral meshes capable to help identify the internal voids of the <b>COVID-19 spike glycoprotein. </b> The middle and right meshes are slices. Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vxx 6vxx], [https://www.rcsb.org/ Protein Data Bank]. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 22:30, 23 July 2020
Contents
3D Example Meshes
The directory containing the 3D input data is located in the 3D folder of Bioninformatics_Data.
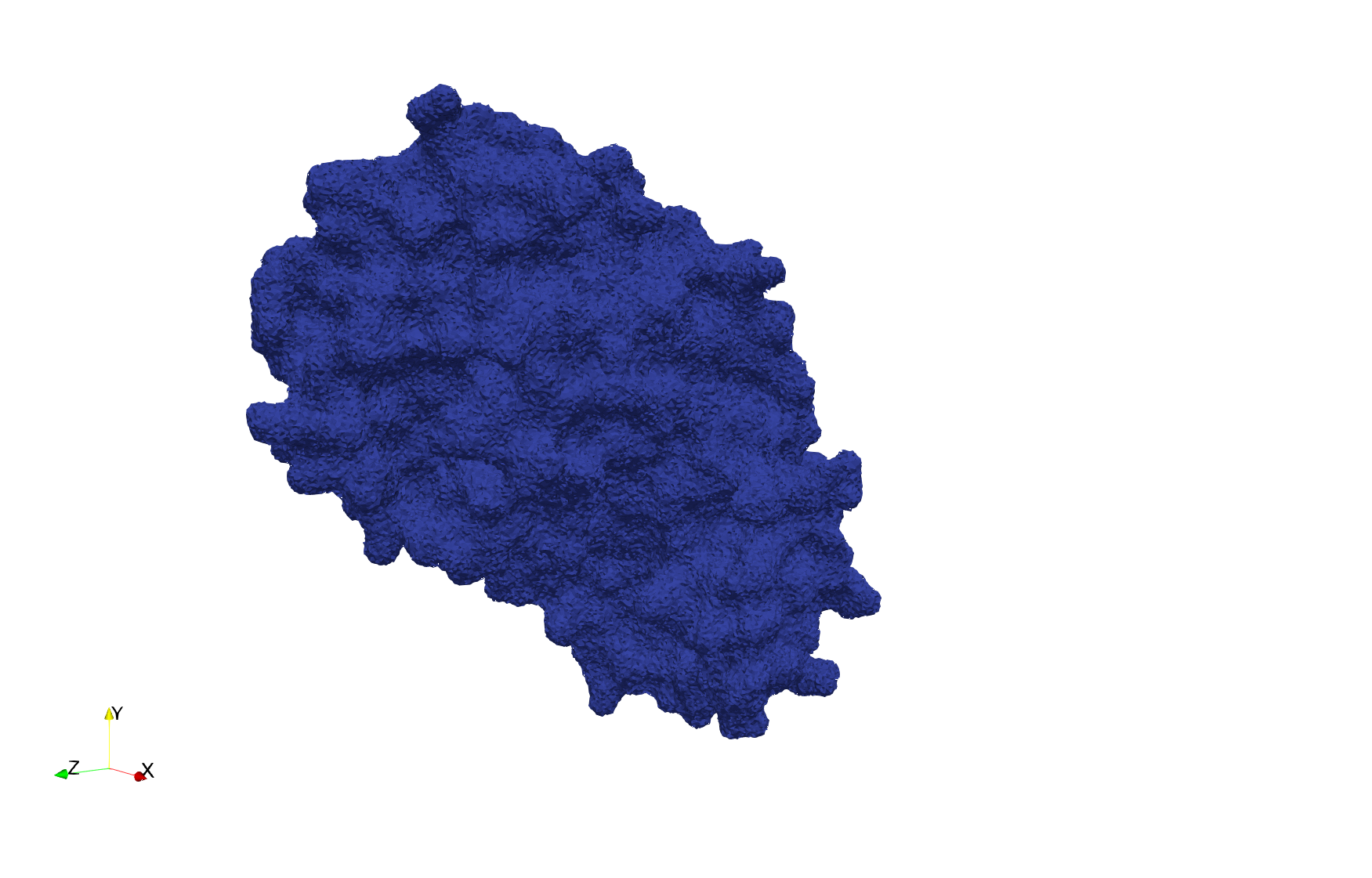
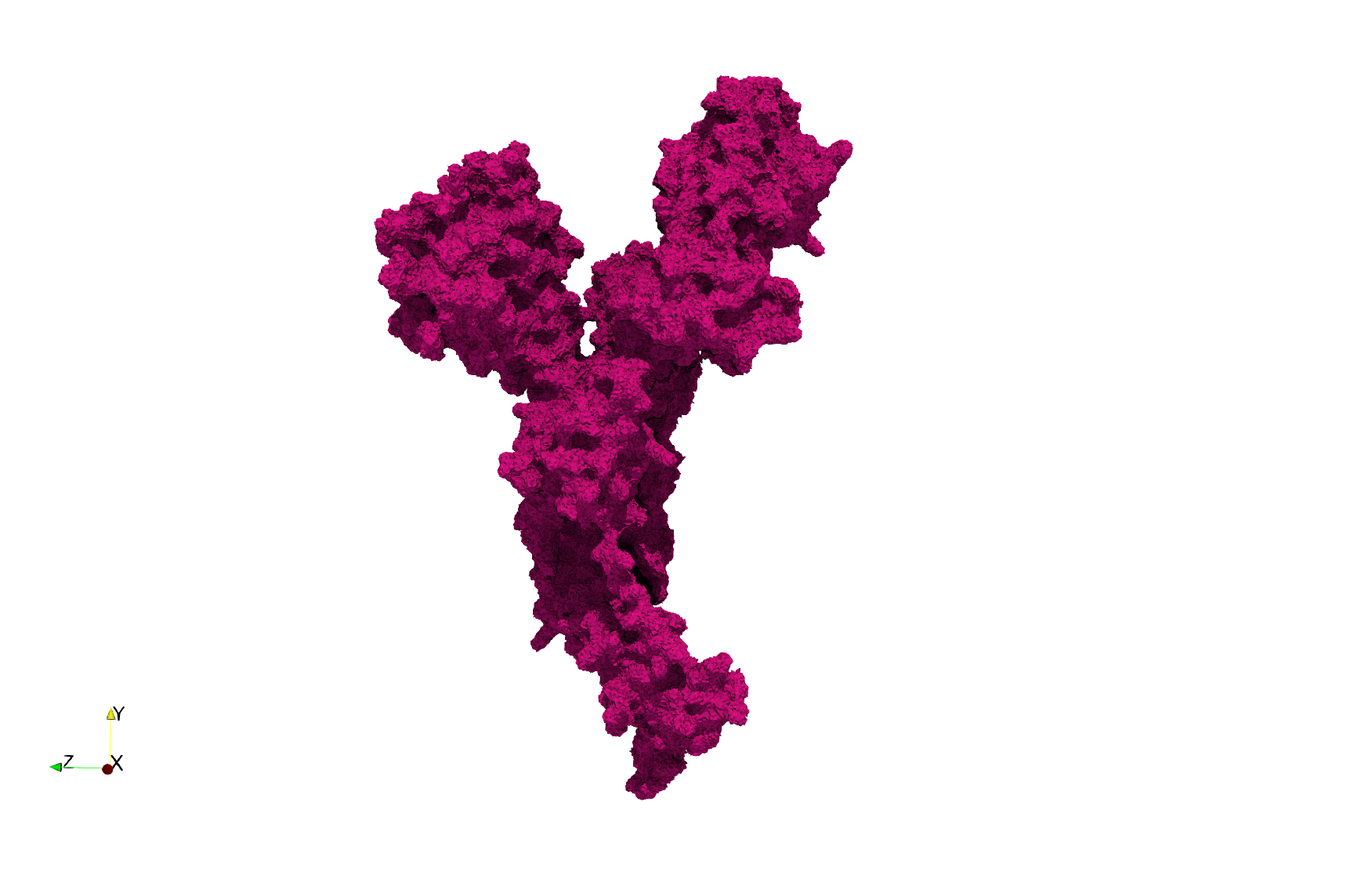
COVID-19-Main-Protease-6y2e
- Input Image
- Input image : Dimensions (284x303x344) with spacing (0.2226907x0.2226907x0.2226907)
- Uniform with Delta = 0.3: 2,509,202 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.3: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Main-Protease-6y2e.nrrd --delta 0.3 --output ./COVID-19-Main-Protease-6y2e,d=0.3.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 Main Protease. Biological assembly data was retrieved from the molecule 6y2e, Protein Data Bank.
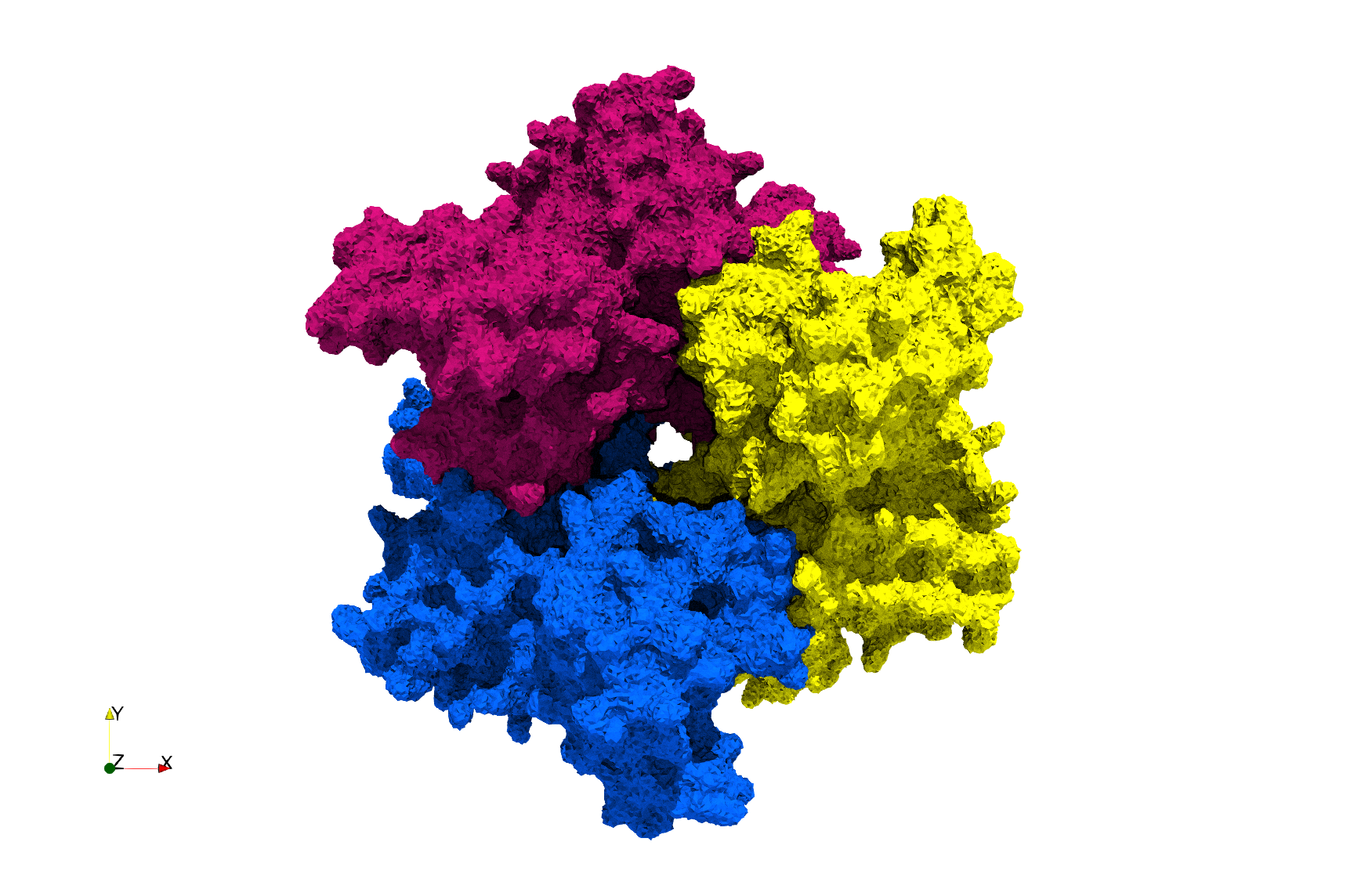
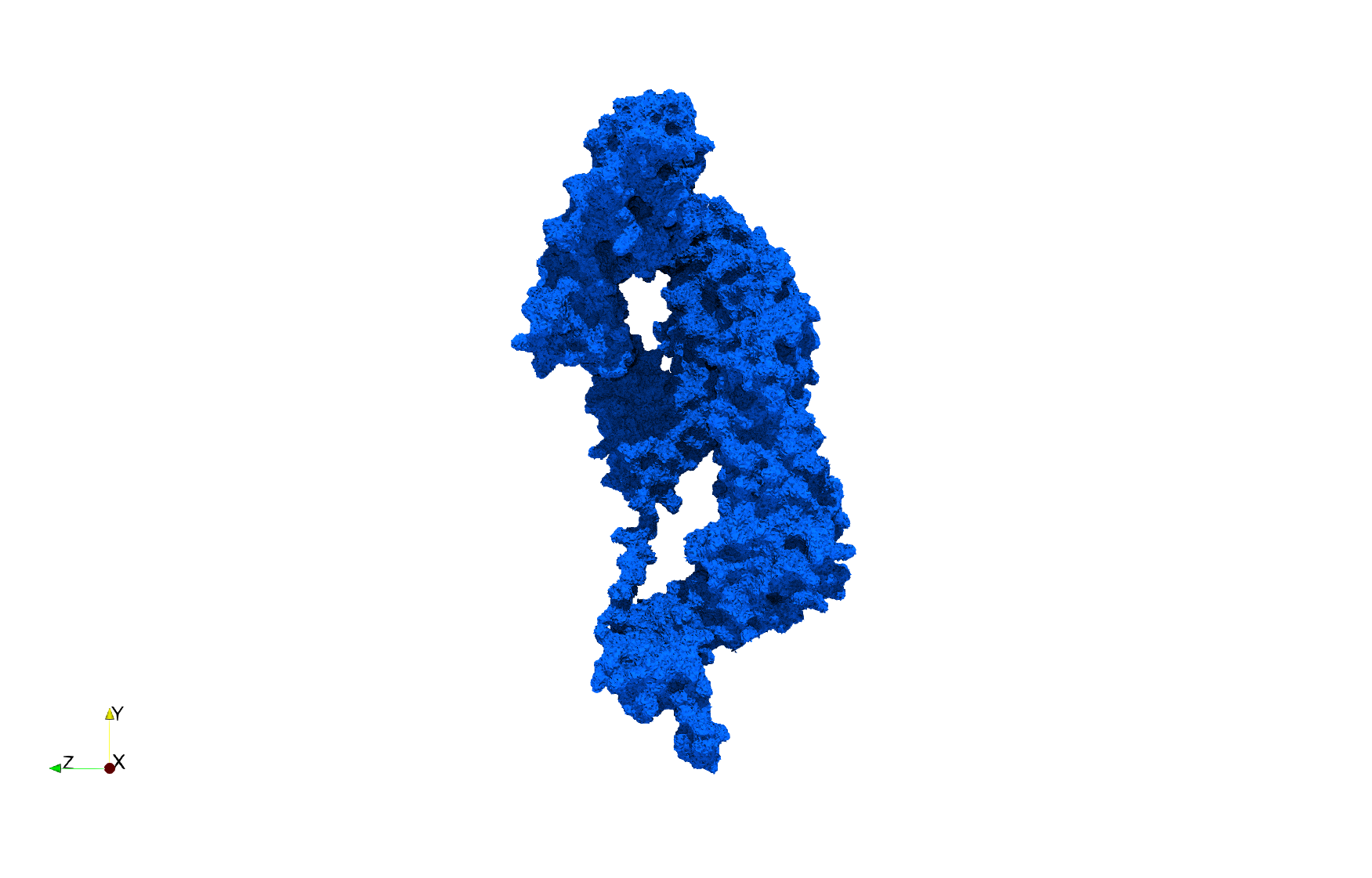
COVID-19-NSP-15-Endoribonuclease-6vww
- [Input Image]
- Input image : Dimensions (303x297x337) with spacing (0.3445713x0.3445713x0.3445713)
- Uniform with Delta = 0.5: 2,310,215 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-NSP-15-Endoribonuclease-6vww.nrrd --delta 0.5 --output ./COVID-19-NSP-15-Endoribonuclease-6vww,d=0.5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 NSP15 Endoribonuclease. Biological assembly data was retrieved from the molecule 6vww, Protein Data Bank.
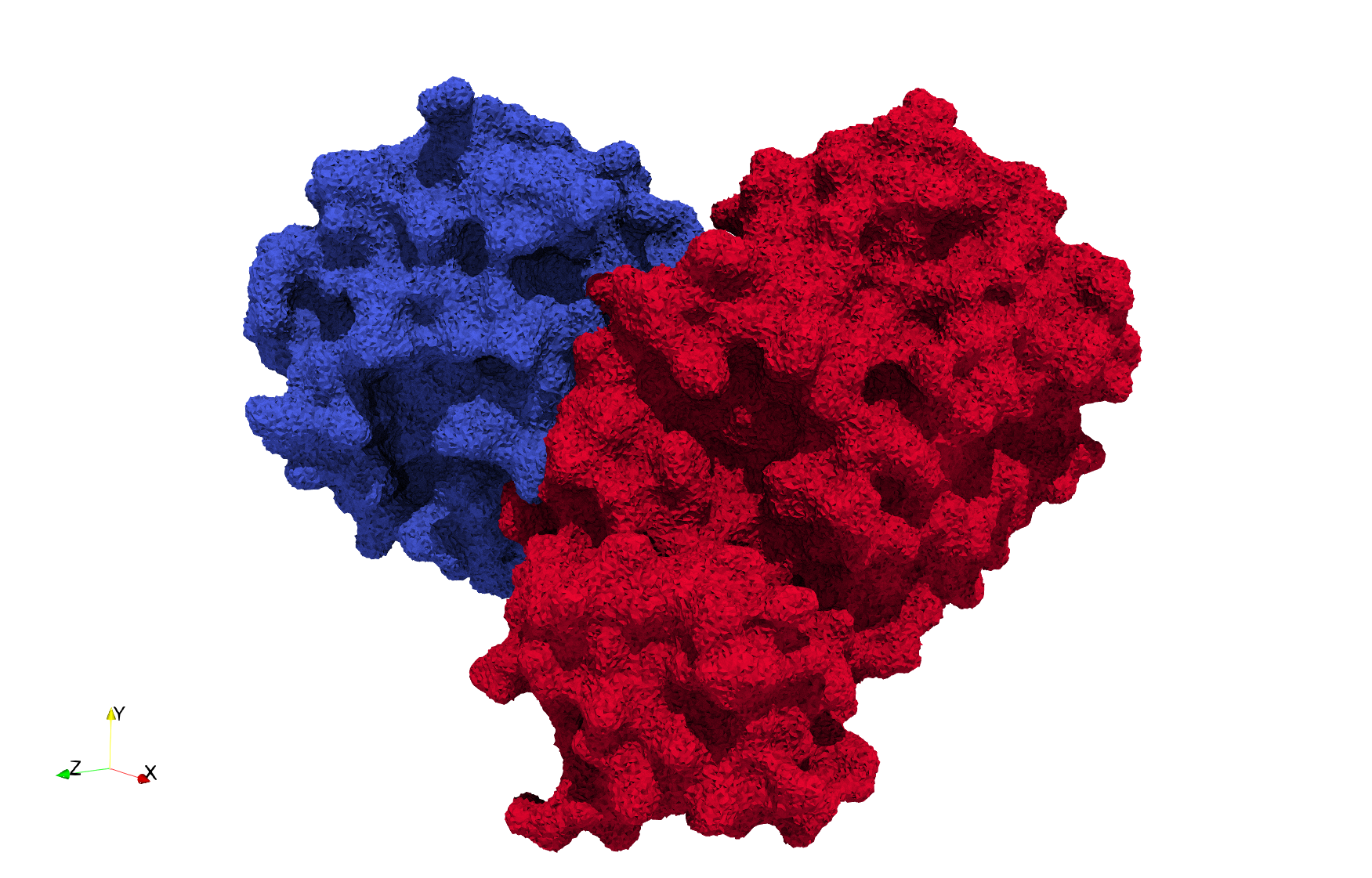
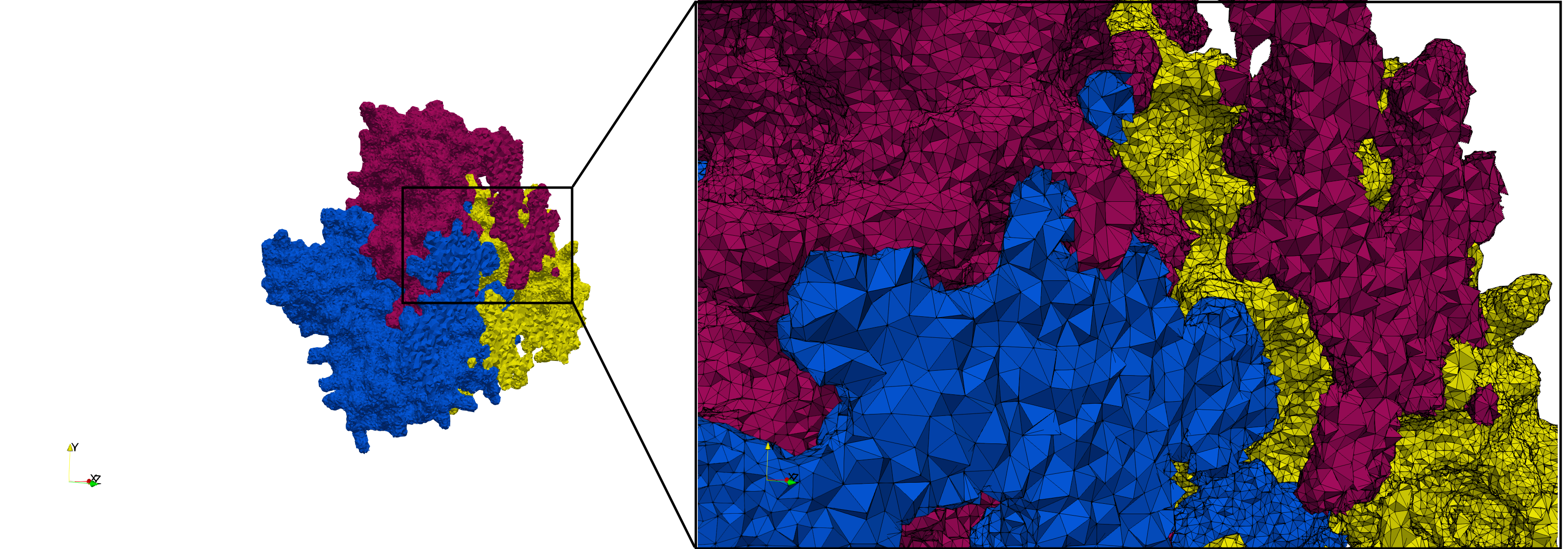
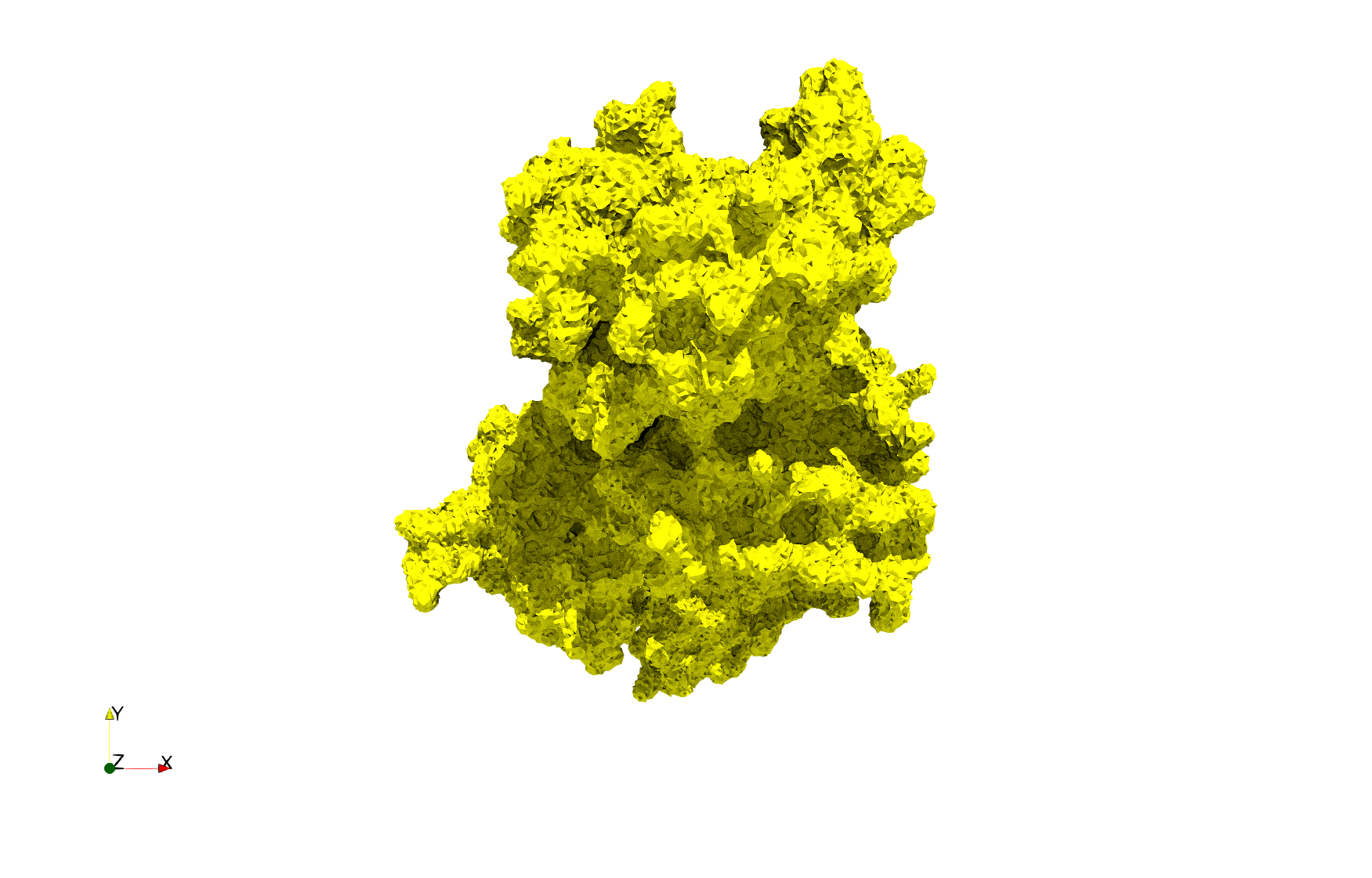
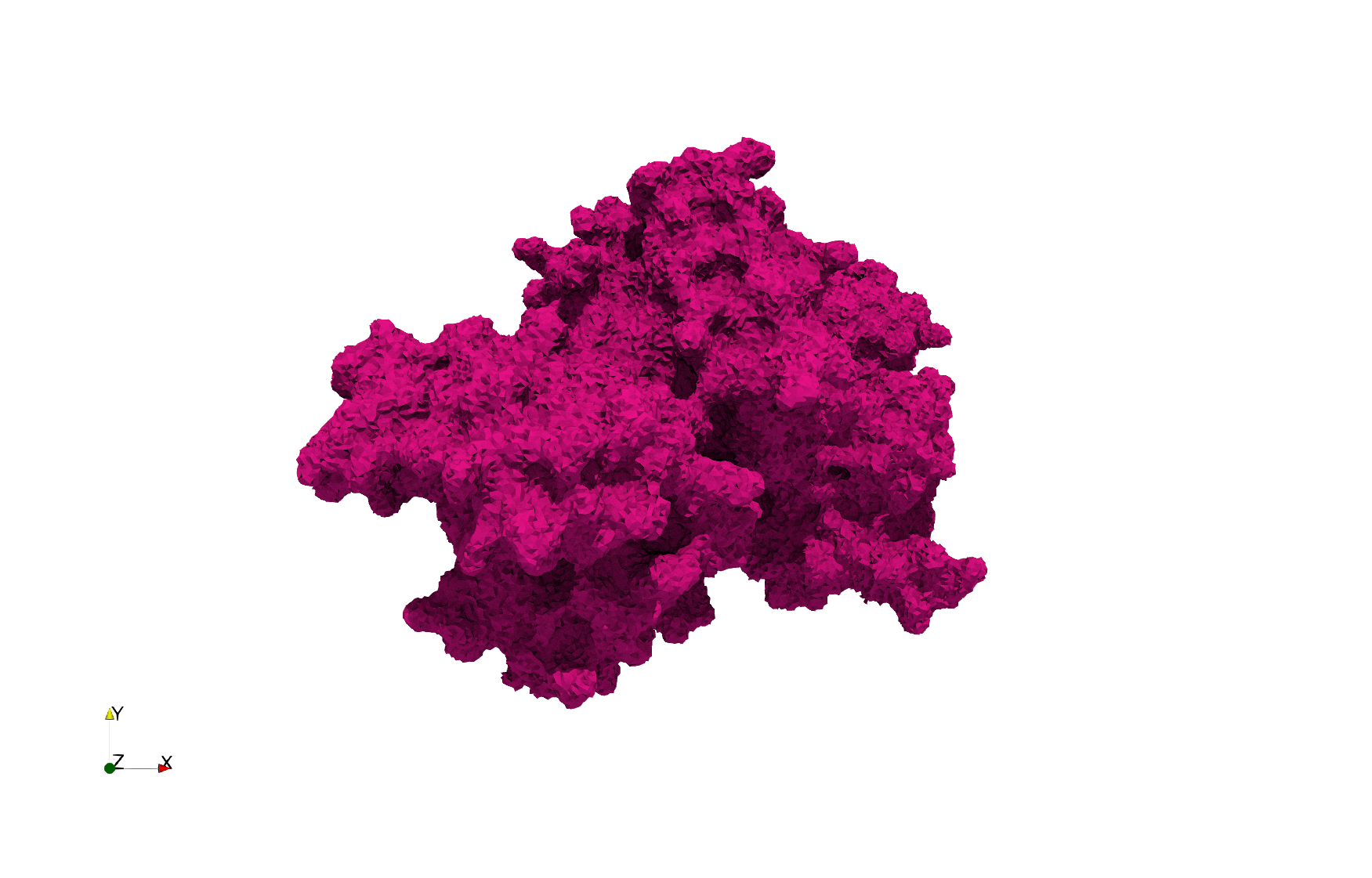
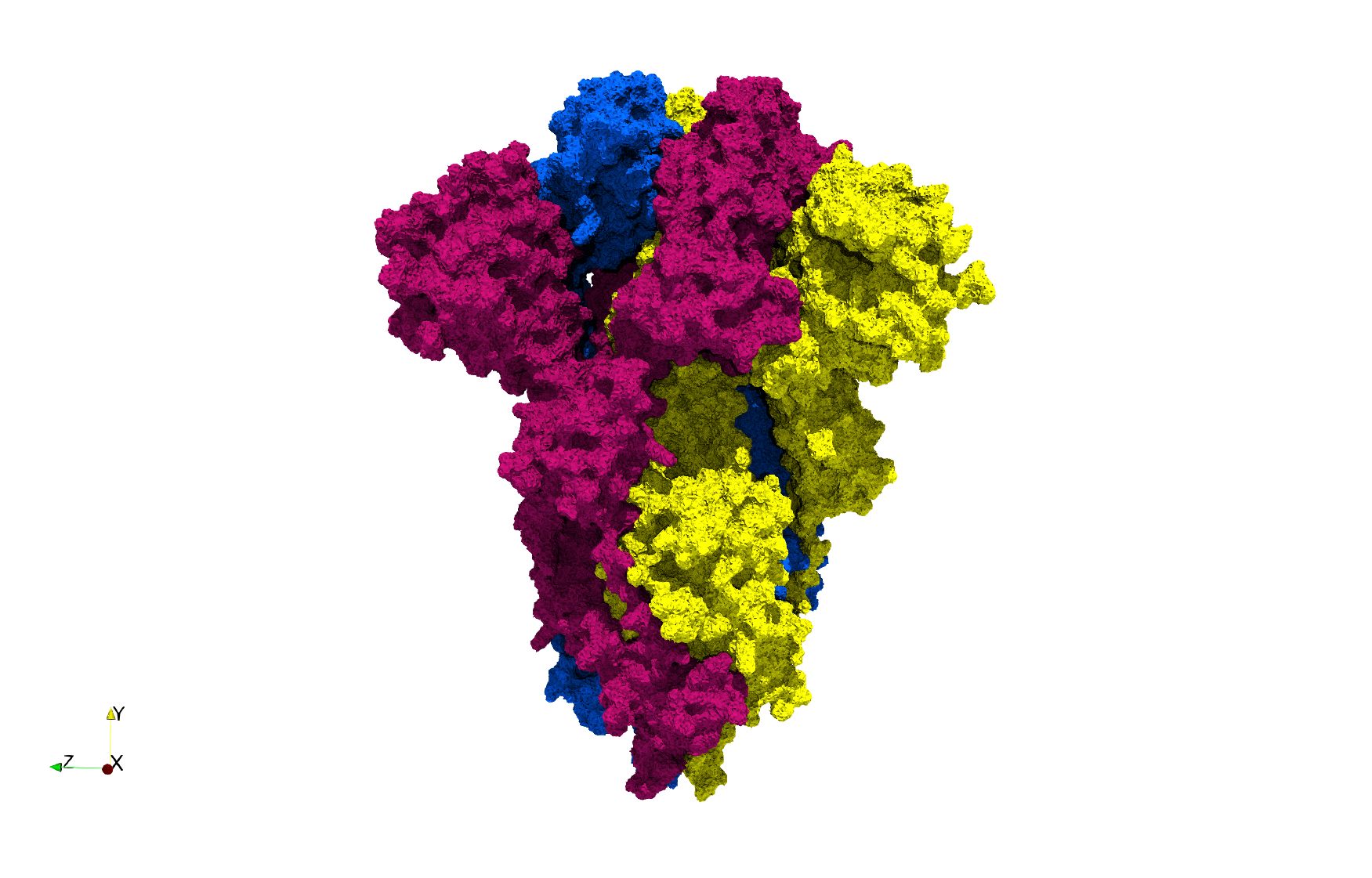
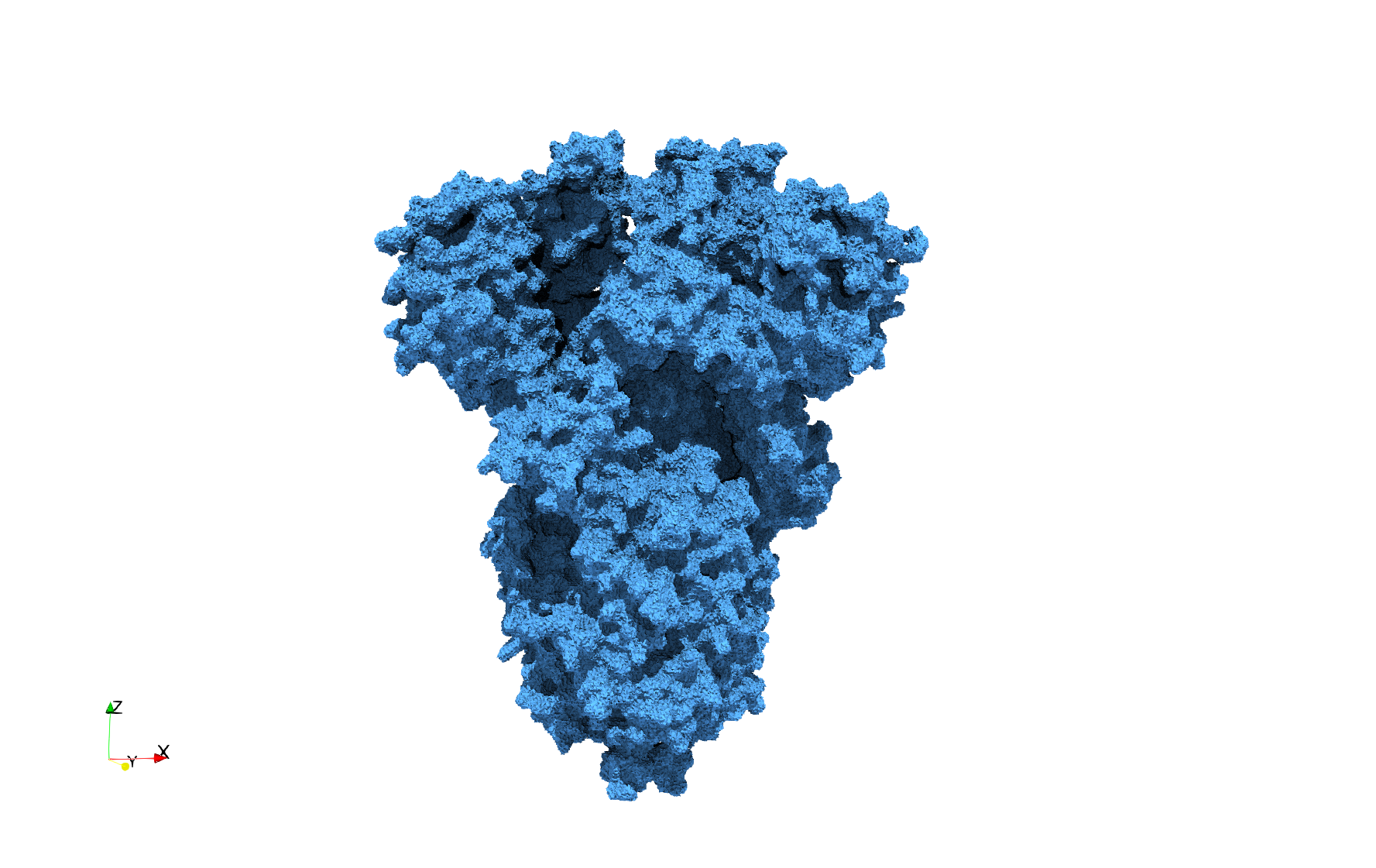
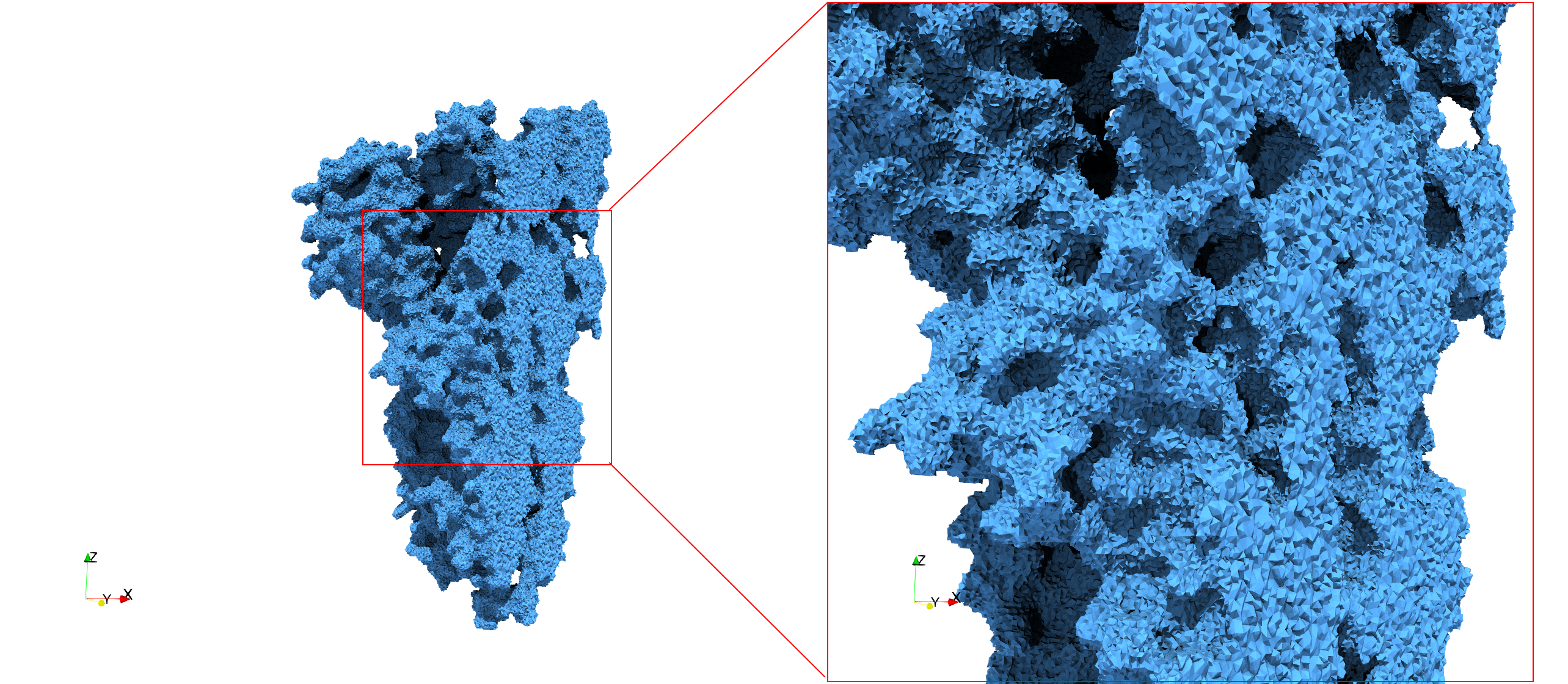
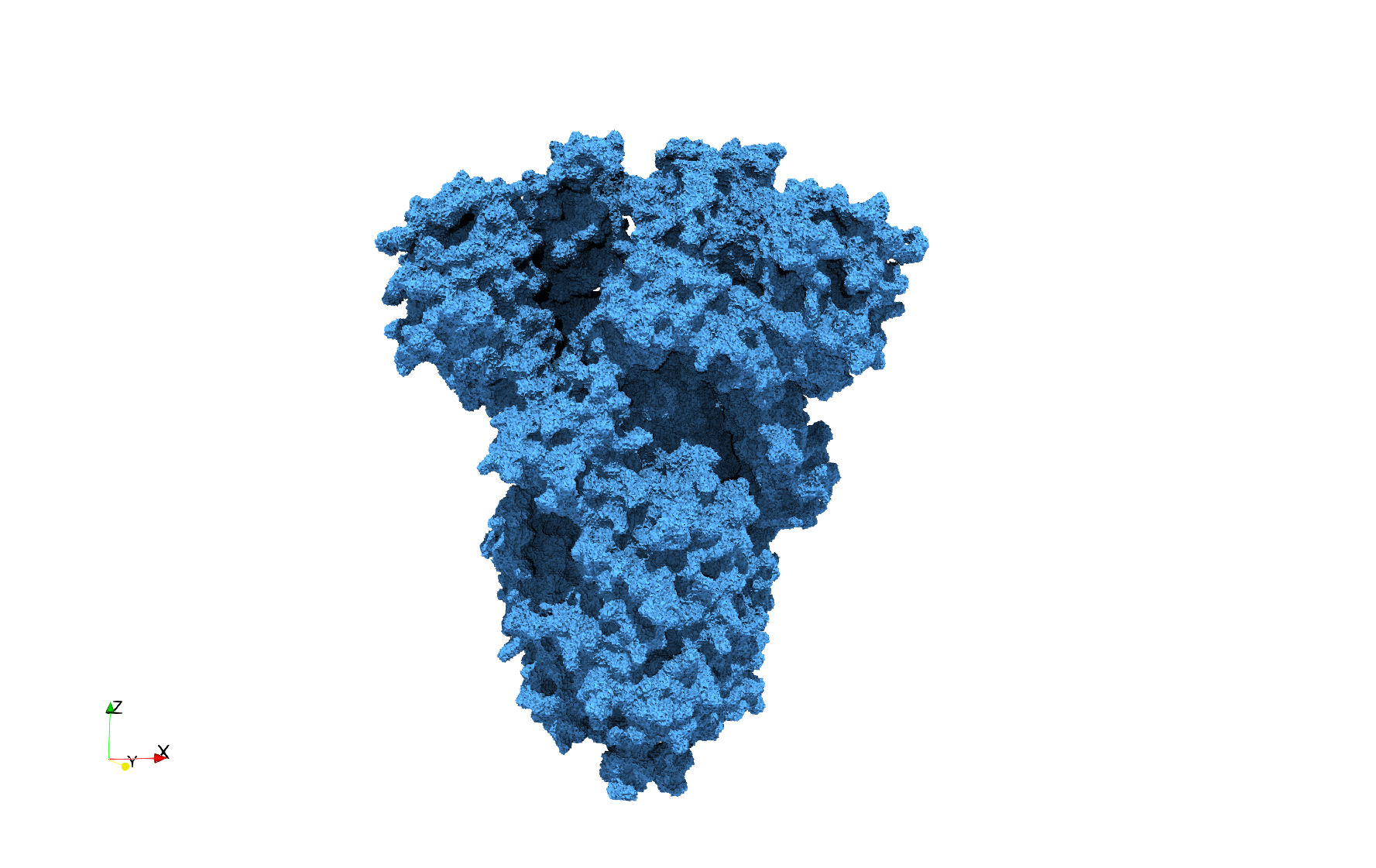
COVID-19-Spike-Glycoprotein-6vxx
- Input Image
- Input image : Dimensions (281x380x302) with spacing (0.427871x0.427871x0.427871)
- Uniform with Delta = 0.5: 3,260,055 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vxx.nrrd --delta 0.5 --output ./COVID-19-Spike-Glycoprotein-6vxx,d=5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 spike glycoprotein. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
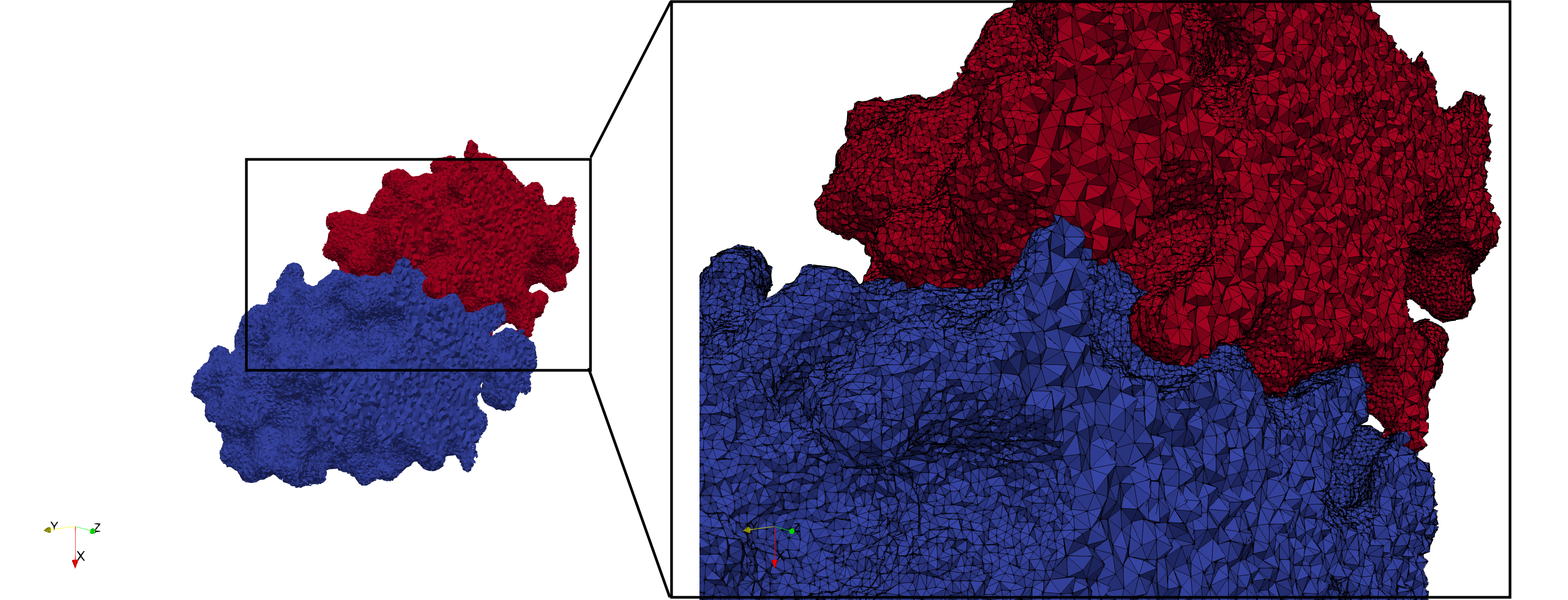
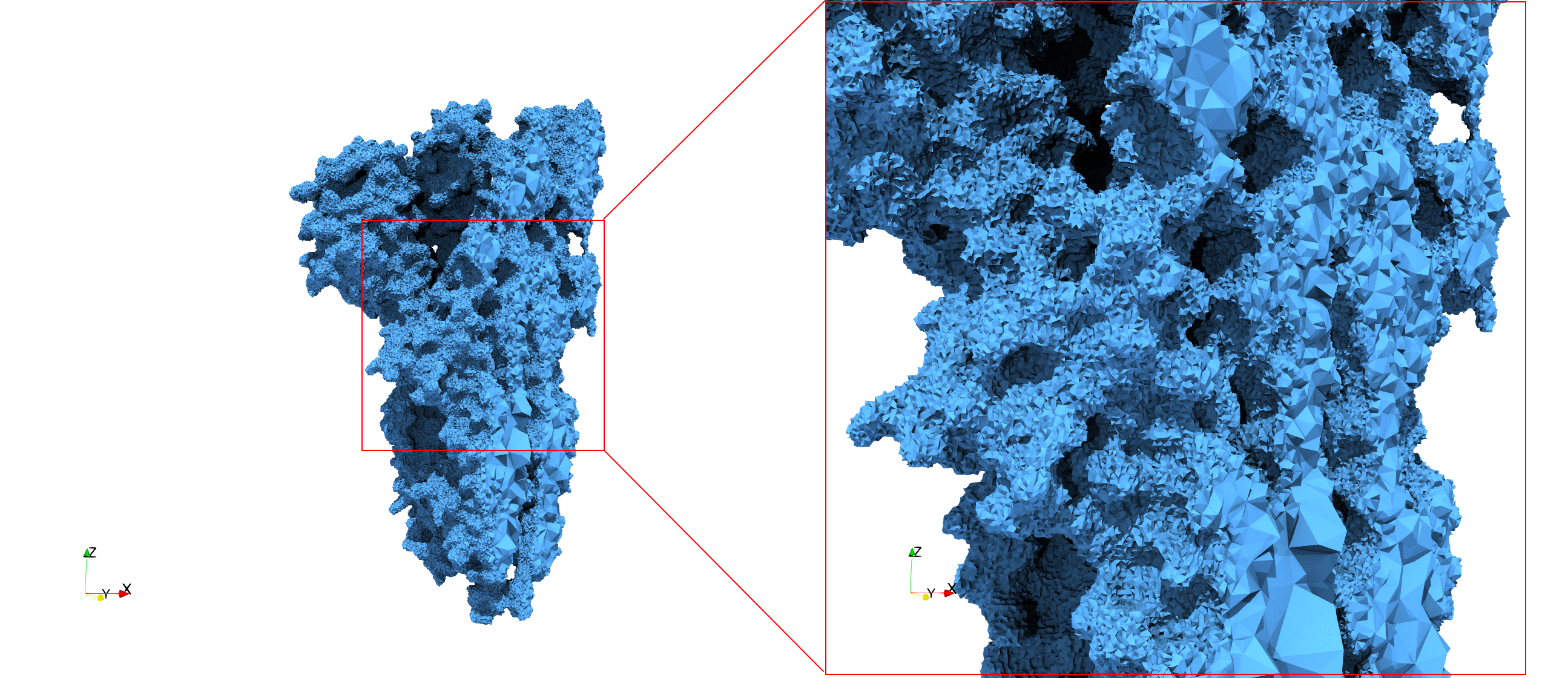
COVID-19-Spike-Glycoprotein-6vsb
- Input Image
- Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042)
- Uniform with Delta = 0.4: 5,731,833 tetrahedra
- Graded with Delta = 0.4: 3,794,223 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4.vtk
Graded with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --volume-grading --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.vtk
Uniform and graded tetrahedral meshes capable to help identify the internal voids of the COVID-19 spike glycoprotein. The middle and right meshes are slices. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.