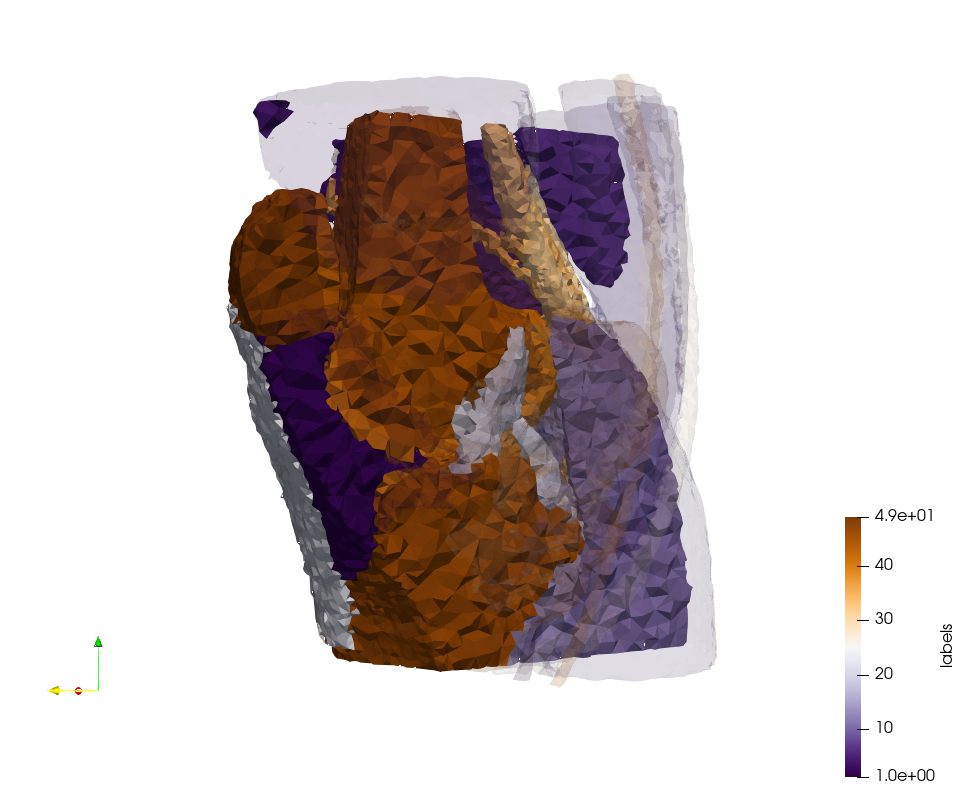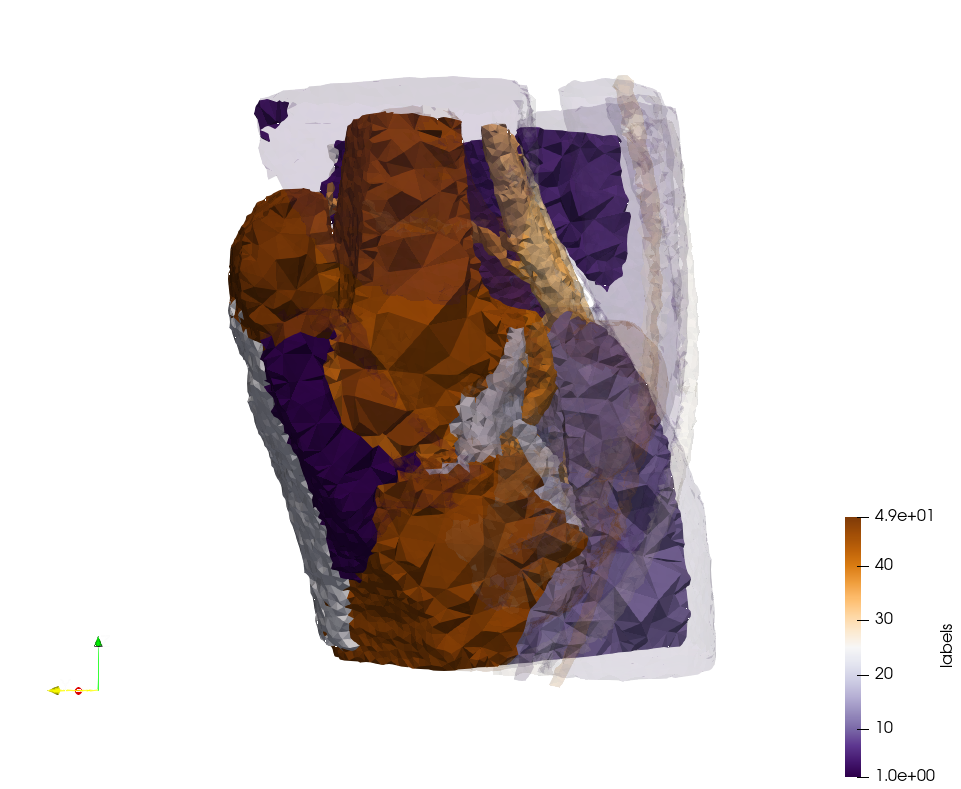Difference between revisions of "Medical Imaging Example Meshes"
From crtc.cs.odu.edu
Spyridon97 (talk | contribs) |
Spyridon97 (talk | contribs) (→Ircad2-Removed-Tissues) |
||
| Line 52: | Line 52: | ||
=Ircad2-Removed-Tissues= | =Ircad2-Removed-Tissues= | ||
| − | * Input image : Dimensions ( | + | * Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002) |
| − | * Uniform case: | + | * Uniform with Delta = 4.92032 (Default) case: 16,685,902 tetrahedra |
| − | * Graded | + | * Graded with Delta = 4.92032 (Default) case: 14,462,494 tetrahedra |
| − | [[File: | + | [[File:Ircad2-Removed-Tissues,d=4.92032.png|500px|Ircad2-Removed-Tissues,d=4.92032.png]] |
| + | [[File:Ircad2-Removed-Tissues,d=4.92032,graded.png|500px|Ircad2-Removed-Tissues,d=4.92032,graded.png]] | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
| − | |||
</gallery> | </gallery> | ||
Commands to generate meshes : | Commands to generate meshes : | ||
| − | ''' | + | '''Uniform :''' [ Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/ | + | docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd --output ./Ircad2-Removed-Tissues,d=4.92032.vtk |
</pre> | </pre> | ||
| − | ''' | + | '''Graded :''' [ Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd -- | + | docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd --volume-grading --output ./Ircad2-Removed-Tissues,d=4.92032,graded.vtk |
</pre> | </pre> | ||
Revision as of 17:45, 26 February 2020
The directory containing the input data is Medical_Imaging_Data.
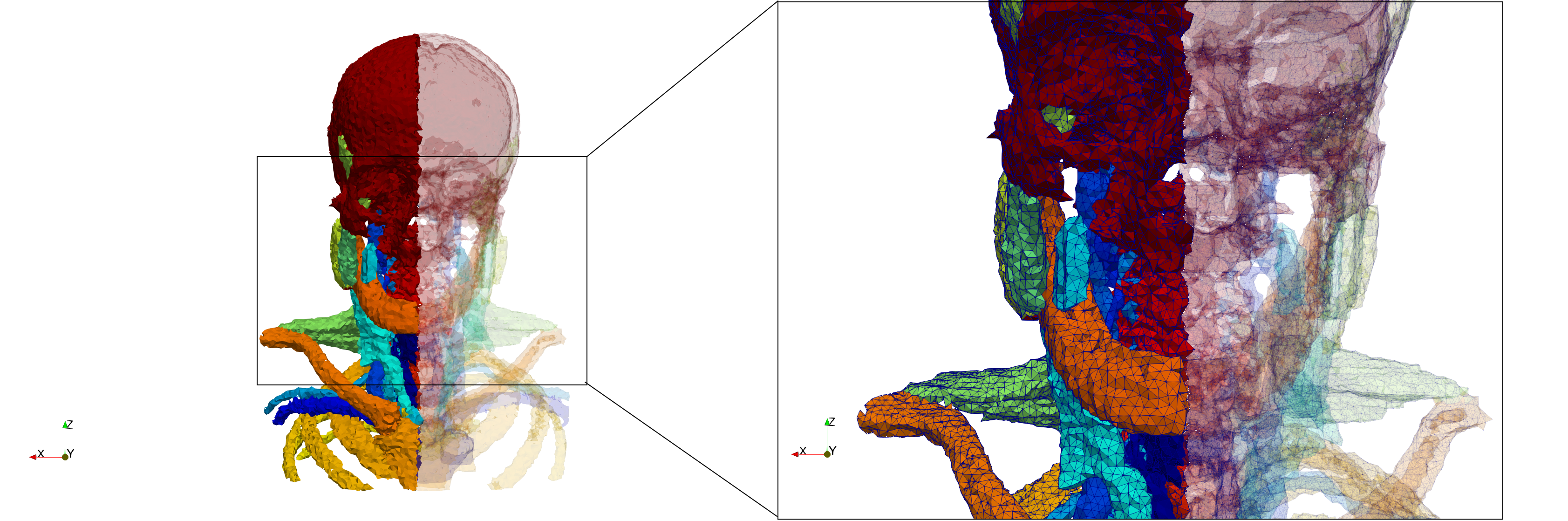
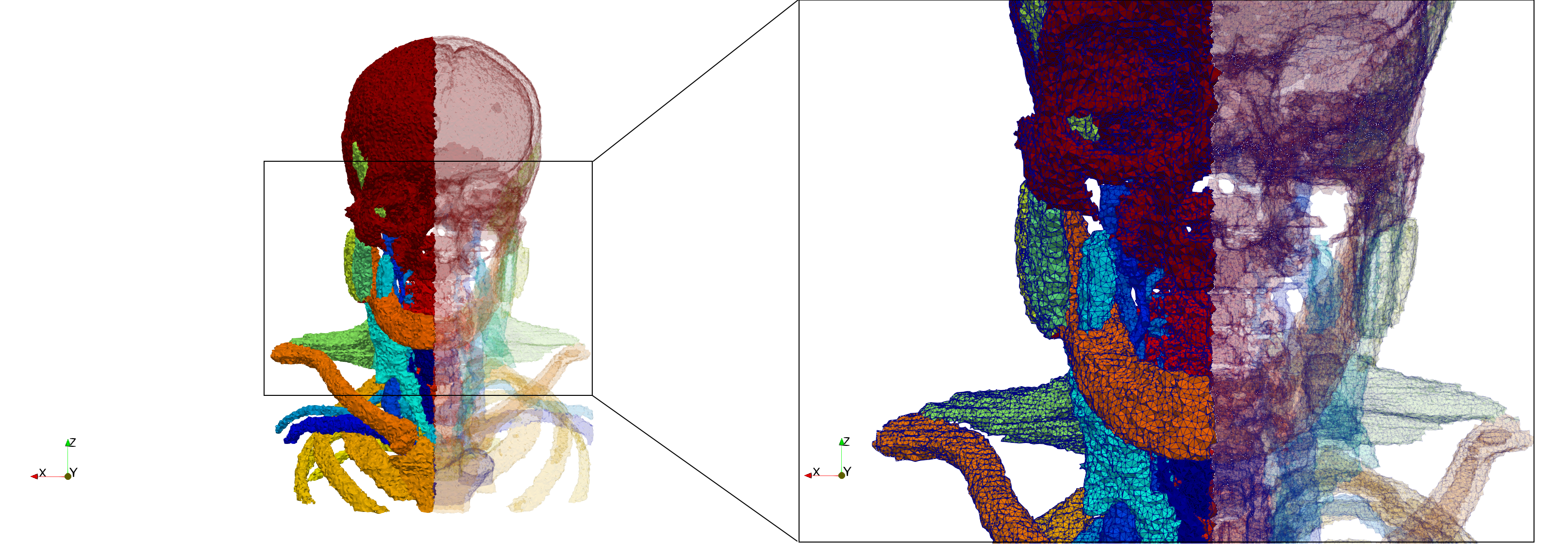
Head-Neck
- Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002)
- Delta = 2.49023 (Default calculated value) case: 205,416 tetrahedra
- Delta = 1.5 case: 770,853 tetrahedra
Commands to generate meshes :
Delta = 2.49023 : [ Output Mesh]
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Head-Neck.mha --output ./Head-Neck,d=2.49023.vtk
Delta = 1.5 : [ Output Mesh]
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
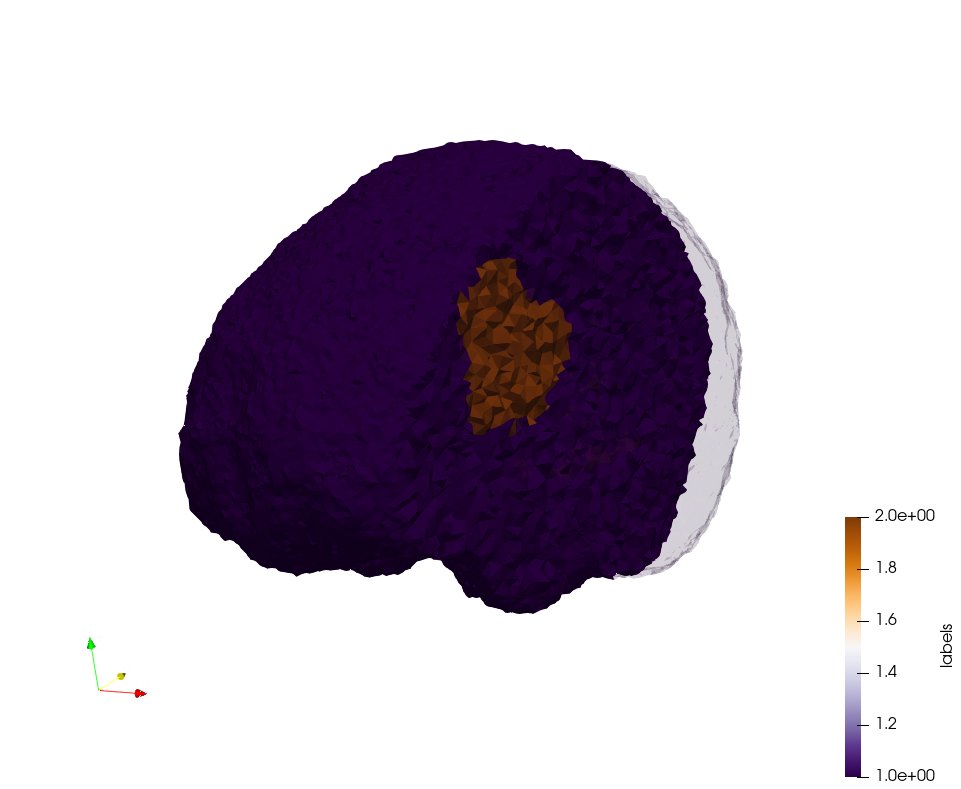
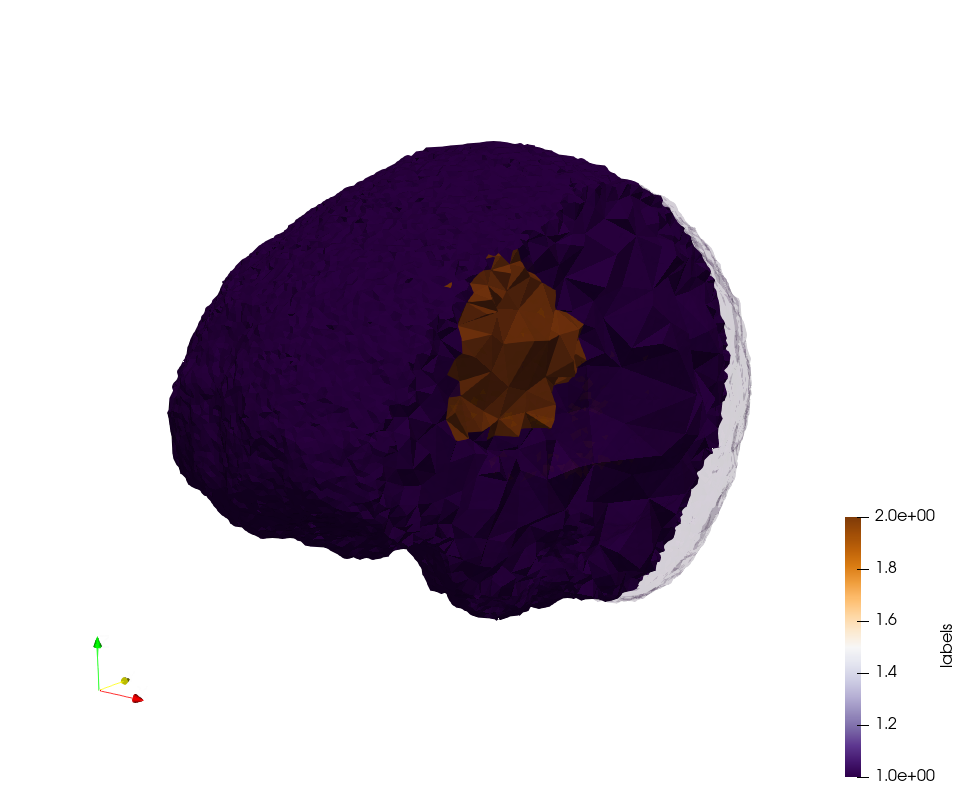
Brain-With-Tumor-Case17
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Brain-With-Tumor-Case17.nii --delta 1.5 --output ./Brain-With-Tumor-Case17,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Brain-With-Tumor-Case17.nii --delta 1.5 --volume-grading --output ./Brain-With-Tumor-Case17,d=1.5,grading.vtk
Knee-Char
Generated mesh with delta = 1,5, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Knee-Char.mha --delta 1.5 --output ./Knee-Char,d=1.5.vtk
Generated mesh with delta = 1.5 and volume-grading, Output Mesh
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Knee-Char.mha --delta 1.5 --volume-grading --output ./Knee-Char,d=1.5,grading.vtk
Ircad2-Removed-Tissues
- Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = 4.92032 (Default) case: 16,685,902 tetrahedra
- Graded with Delta = 4.92032 (Default) case: 14,462,494 tetrahedra
Ircad2-Removed-Tissues,d=4.92032.png Ircad2-Removed-Tissues,d=4.92032,graded.png
Commands to generate meshes :
Uniform : [ Output Mesh]
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd --output ./Ircad2-Removed-Tissues,d=4.92032.vtk
Graded : [ Output Mesh]
docker run -v $(pwd):/data/ crtc_i2m podm3d --input ./Medical_Imaging_Data/Ircad2-Removed-Tissues.nrrd --volume-grading --output ./Ircad2-Removed-Tissues,d=4.92032,graded.vtk