Difference between revisions of "Medical Imaging Example Meshes"
Spyridon97 (talk | contribs) (→Ircad2) |
Spyridon97 (talk | contribs) (→COVID-19-NSP-15-Endoribonuclease-6vww) |
||
| (39 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
The directory containing the 3D input data is located in the 3D folder of [https://odu.box.com/s/olefferrnksu2nmerbfvbvsz2u4abbdw Medical_Imaging_Data]. | The directory containing the 3D input data is located in the 3D folder of [https://odu.box.com/s/olefferrnksu2nmerbfvbvsz2u4abbdw Medical_Imaging_Data]. | ||
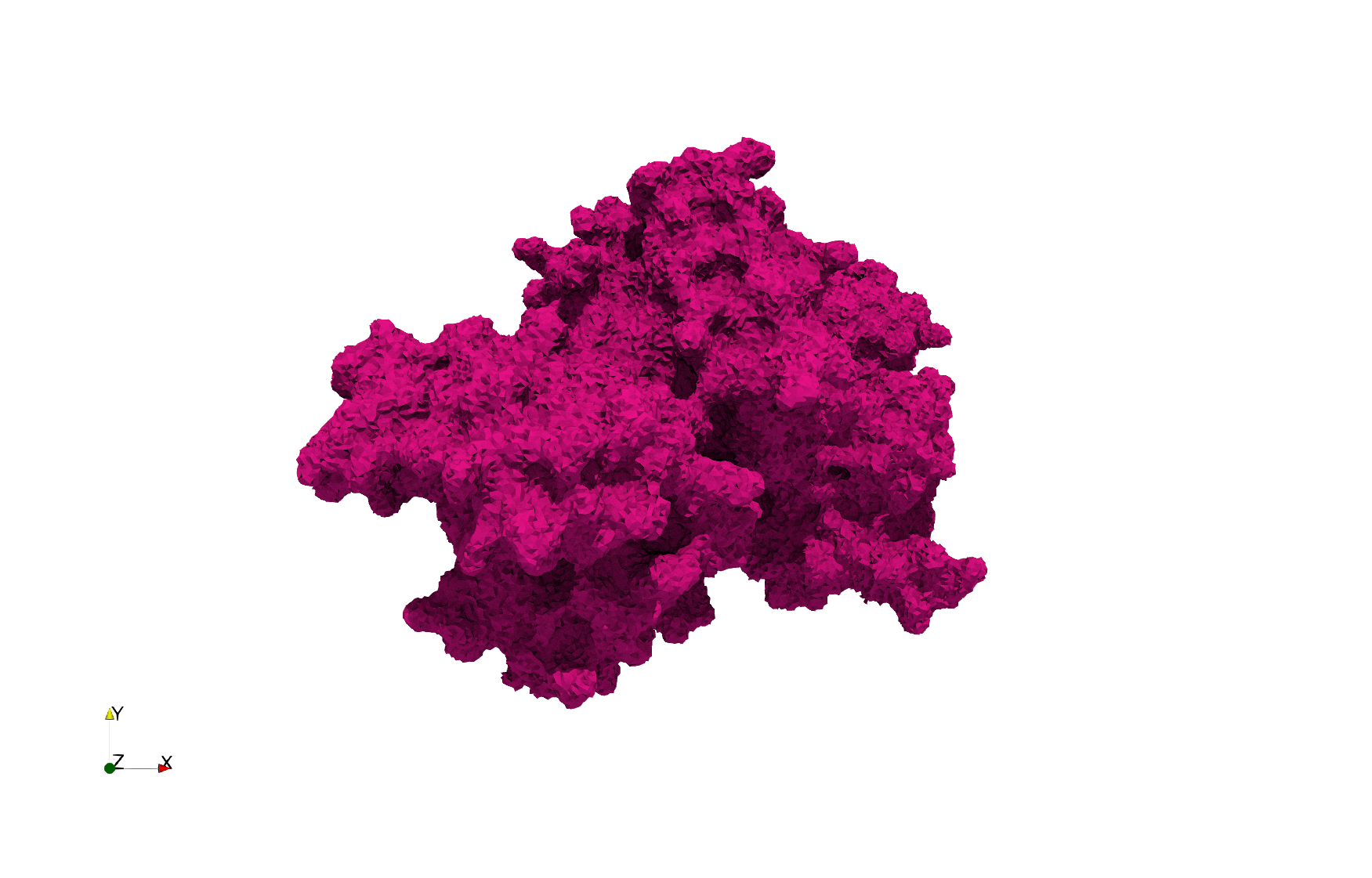
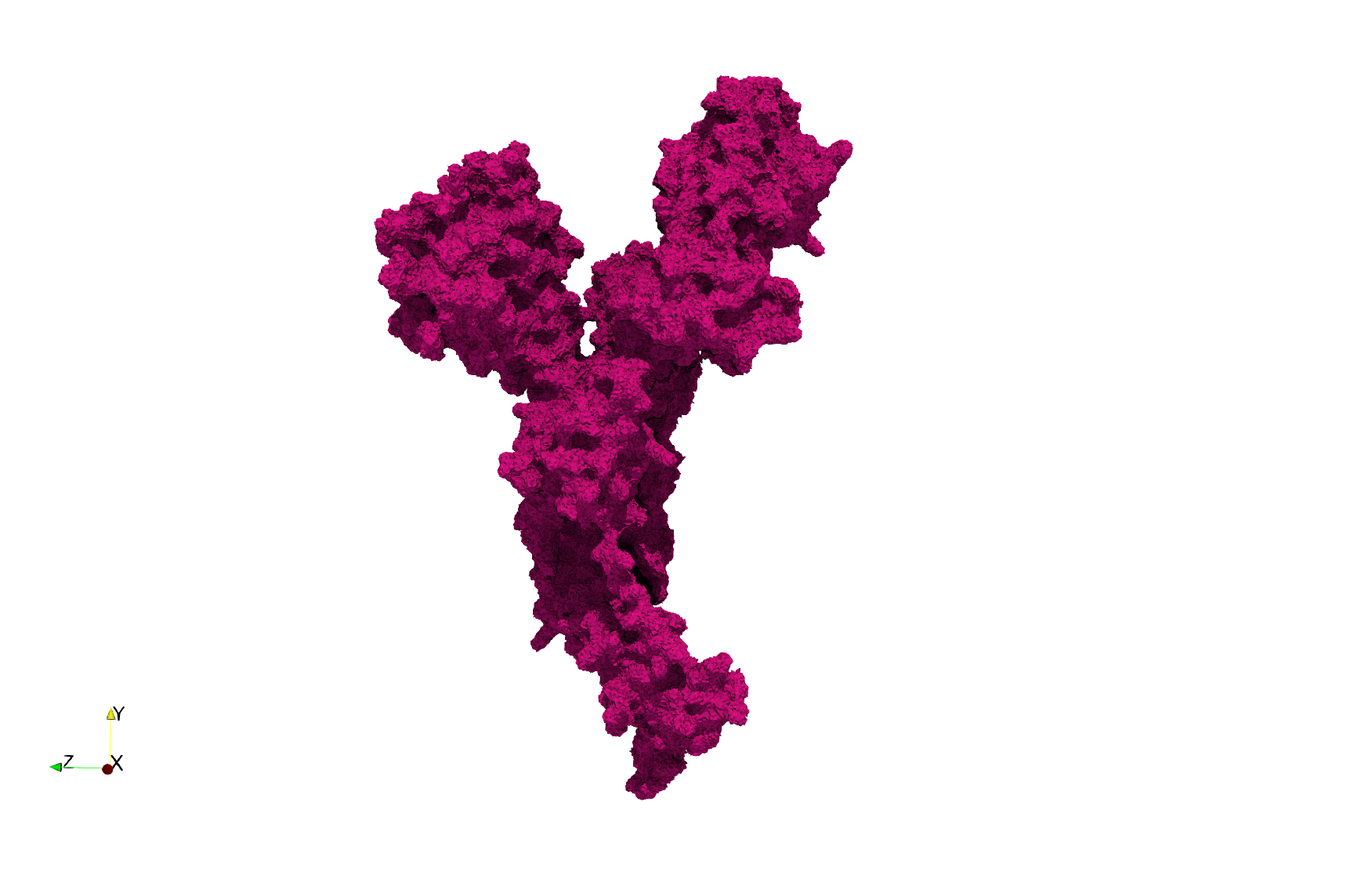
| − | ==COVID-19- | + | ==COVID-19-Main-Protease-6y2e== |
| − | * Input image : Dimensions ( | + | * [https://odu.box.com/s/eb9s3vrvml0gop7mdpwckh6xnn27eiut Input Image] |
| − | * Uniform with Delta = 0. | + | * Input image : Dimensions (284x303x344) with spacing (0.2226907x0.2226907x0.2226907) |
| + | * Uniform with Delta = 0.3: 2,509,202 tetrahedra | ||
<gallery mode="packed" heights=250px> | <gallery mode="packed" heights=250px> | ||
| − | File:COVID-19- | + | File:COVID-19-Main-Protease-6y2e,d=0.5.png |
| − | File:COVID-19- | + | File:COVID-19-Main-Protease-6y2e,d=0.5,clip.png |
| − | File:COVID-19- | + | </gallery> |
| − | File:COVID-19- | + | <gallery mode="packed" heights=250px> |
| + | File:COVID-19-Main-Protease-6y2e,d=0.5,label=1.png | ||
| + | File:COVID-19-Main-Protease-6y2e,d=0.5,label=2.png | ||
</gallery> | </gallery> | ||
Commands to generate meshes: | Commands to generate meshes: | ||
| − | '''Uniform with Delta = 0. | + | '''Uniform with Delta = 0.3:''' [https://odu.box.com/s/jeks38sn5slw38gbit6t0rf25fzv6jpb Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19- | + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Main-Protease-6y2e.nrrd --delta 0.3 --output ./COVID-19-Main-Protease-6y2e,d=0.3.vtk |
</pre> | </pre> | ||
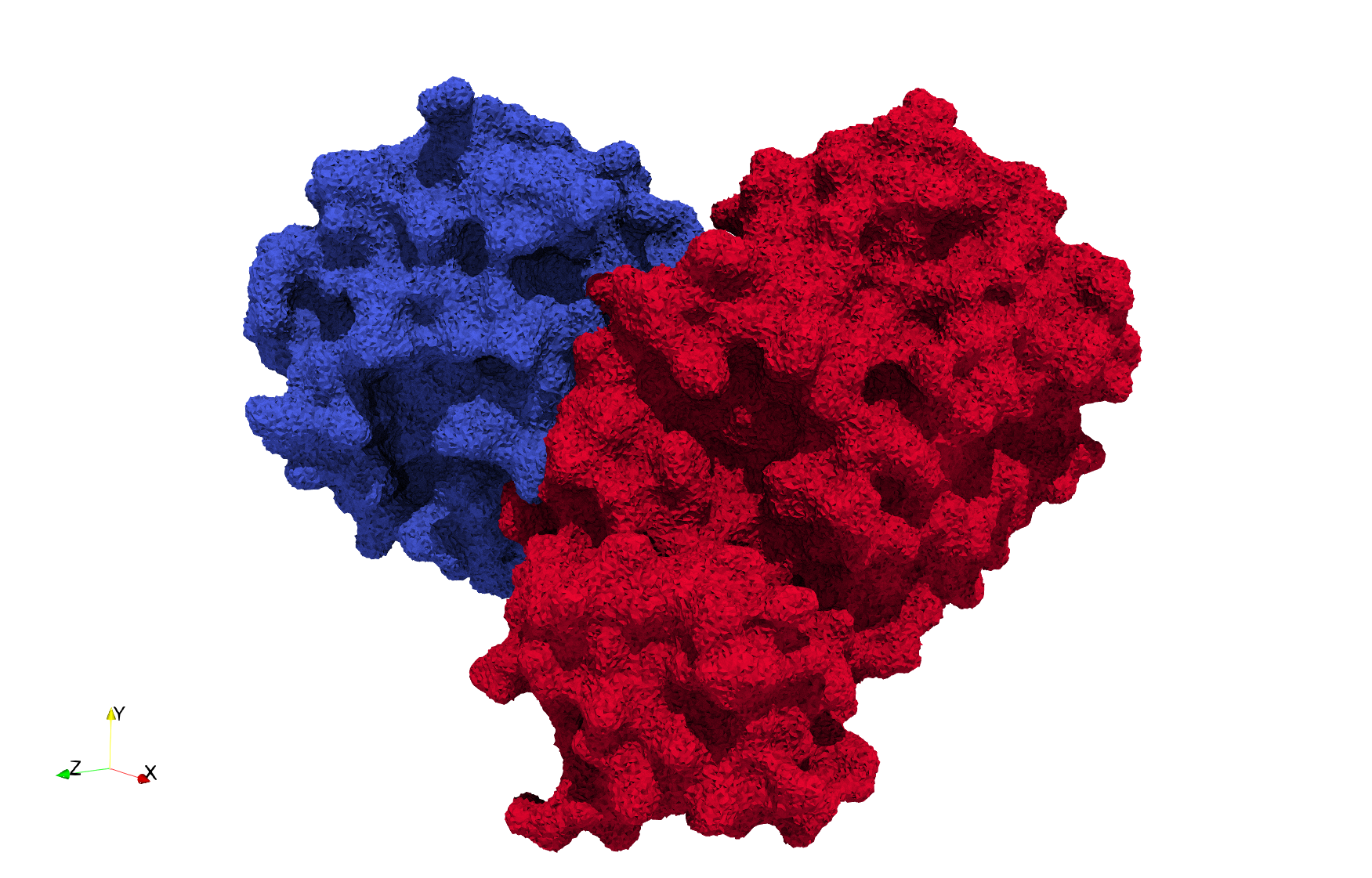
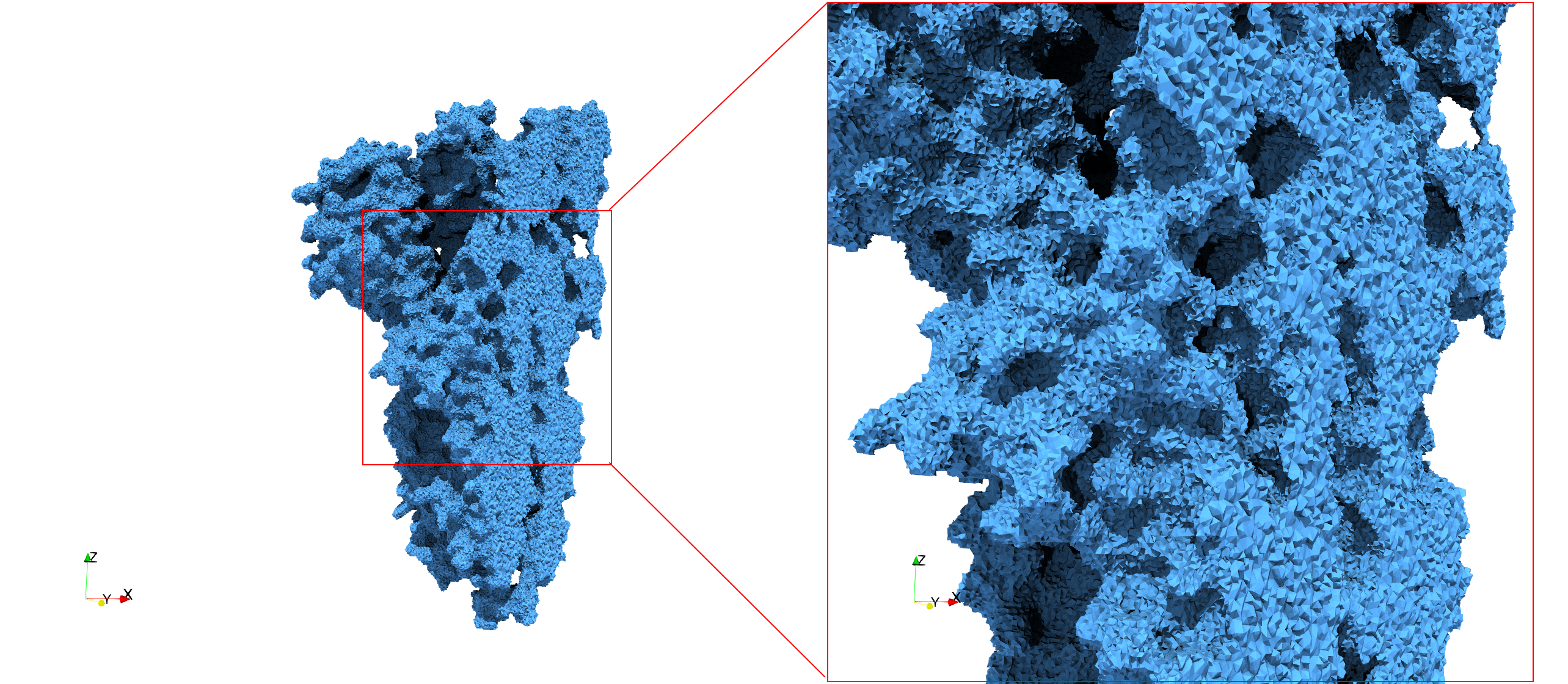
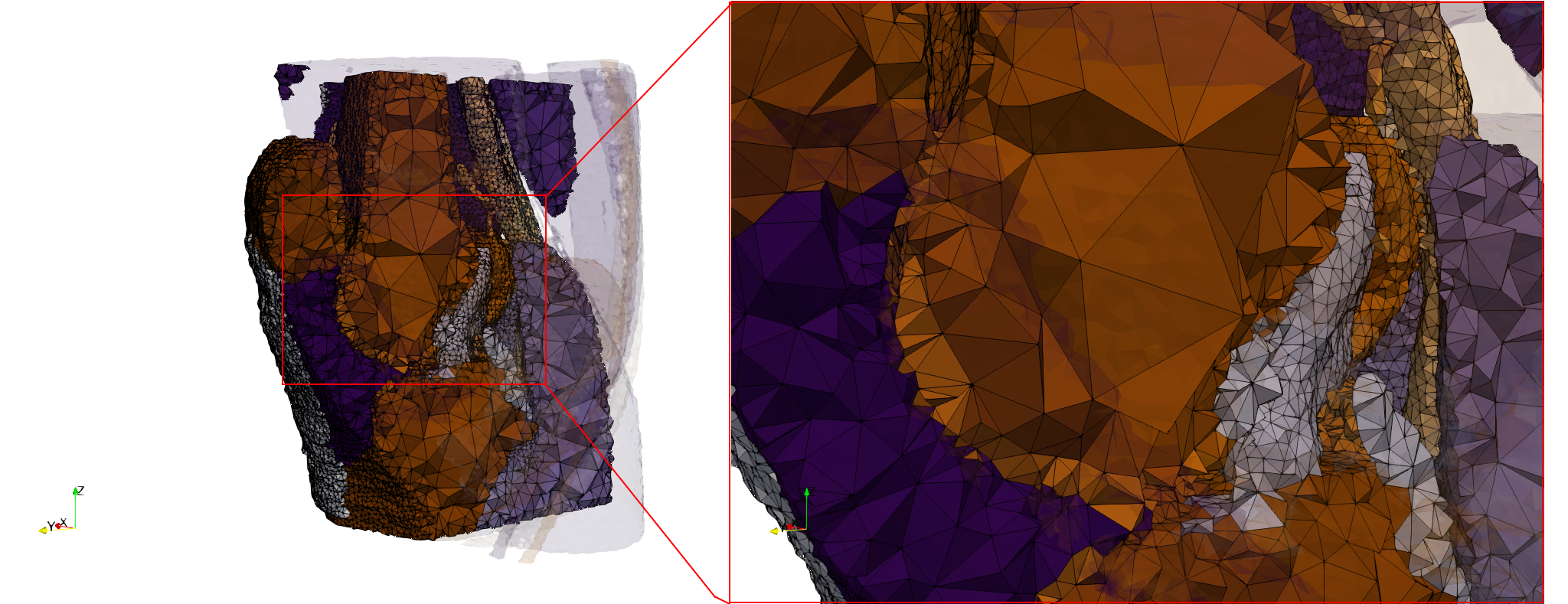
| − | Uniform tetrahedral mesh capable to help identify the internal voids of the <b>COVID-19 | + | Uniform tetrahedral mesh capable to help identify the internal voids of the <b>COVID-19 Main Protease. </b> Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6y2e 6y2e], [https://www.rcsb.org/ Protein Data Bank]. |
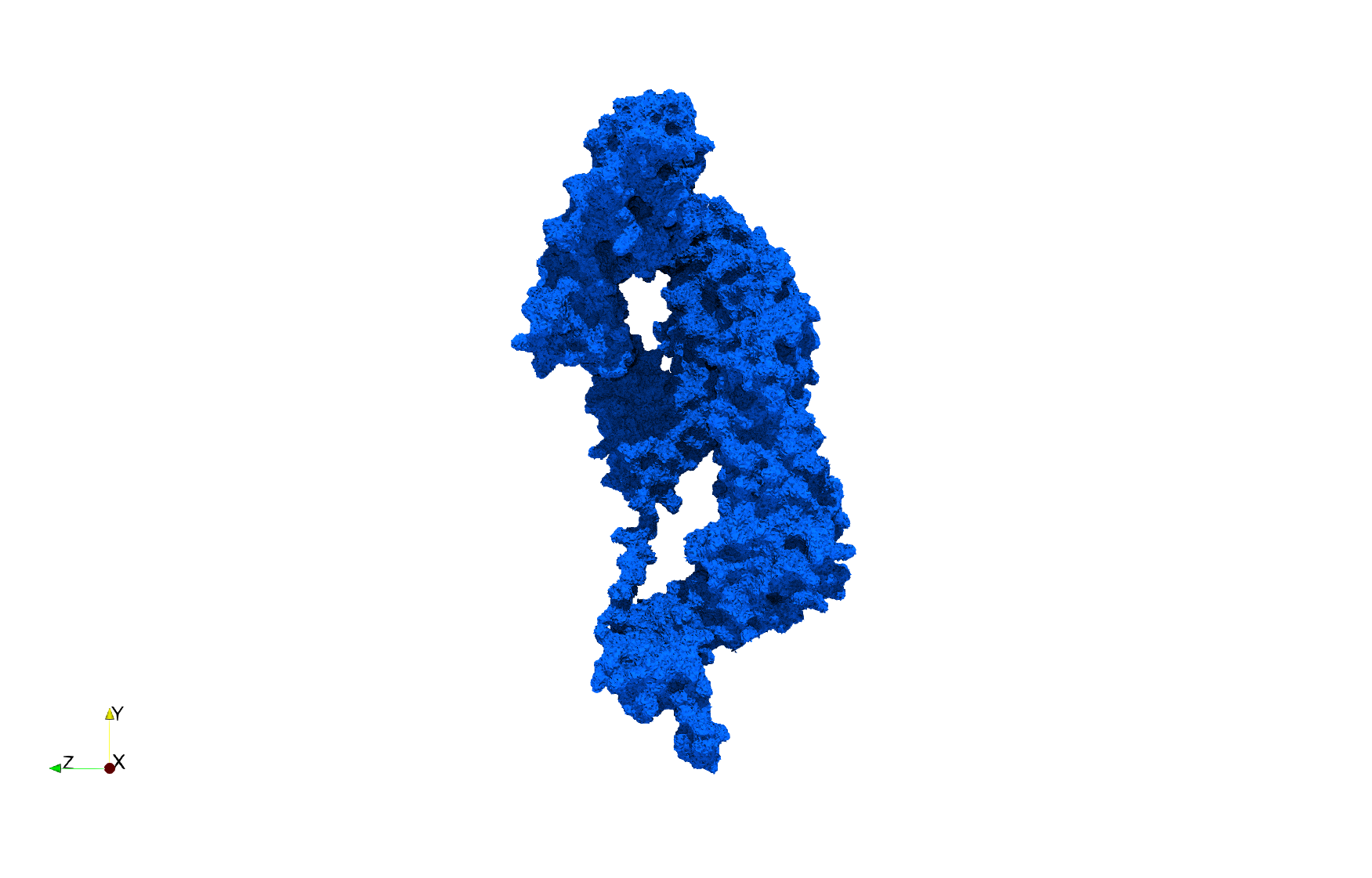
==COVID-19-NSP-15-Endoribonuclease-6vww== | ==COVID-19-NSP-15-Endoribonuclease-6vww== | ||
| + | * [Input Image] | ||
* Input image : Dimensions (303x297x337) with spacing (0.3445713x0.3445713x0.3445713) | * Input image : Dimensions (303x297x337) with spacing (0.3445713x0.3445713x0.3445713) | ||
| − | * Uniform with Delta = 0. | + | * Uniform with Delta = 0.5: 2,310,215 tetrahedra |
<gallery mode="packed" heights=250px> | <gallery mode="packed" heights=250px> | ||
File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3.png | File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3.png | ||
| + | File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3,clip.png | ||
| + | </gallery> | ||
| + | <gallery mode="packed" heights=250px> | ||
File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3,label=1.png | File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3,label=1.png | ||
File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3,label=2.png | File:COVID-19-NSP-15-Endoribonuclease-6vww,d=0.3,label=2.png | ||
| Line 37: | Line 44: | ||
Commands to generate meshes: | Commands to generate meshes: | ||
| − | '''Uniform with Delta = 0. | + | '''Uniform with Delta = 0.5:''' [https://odu.box.com/s/tkk8f8o1tctf52bop07a6hxy87ugt8kg Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-NSP-15-Endoribonuclease-6vww.nrrd --delta 0. | + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-NSP-15-Endoribonuclease-6vww.nrrd --delta 0.5 --output ./COVID-19-NSP-15-Endoribonuclease-6vww,d=0.5.vtk |
</pre> | </pre> | ||
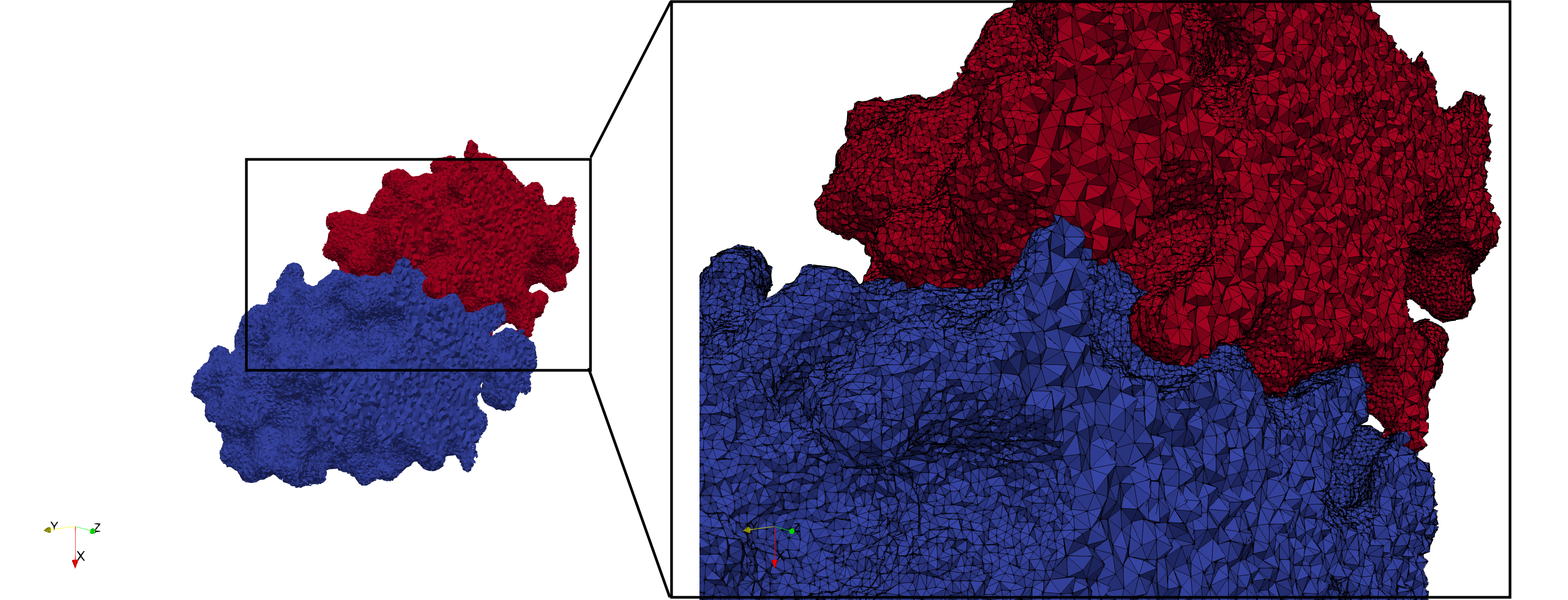
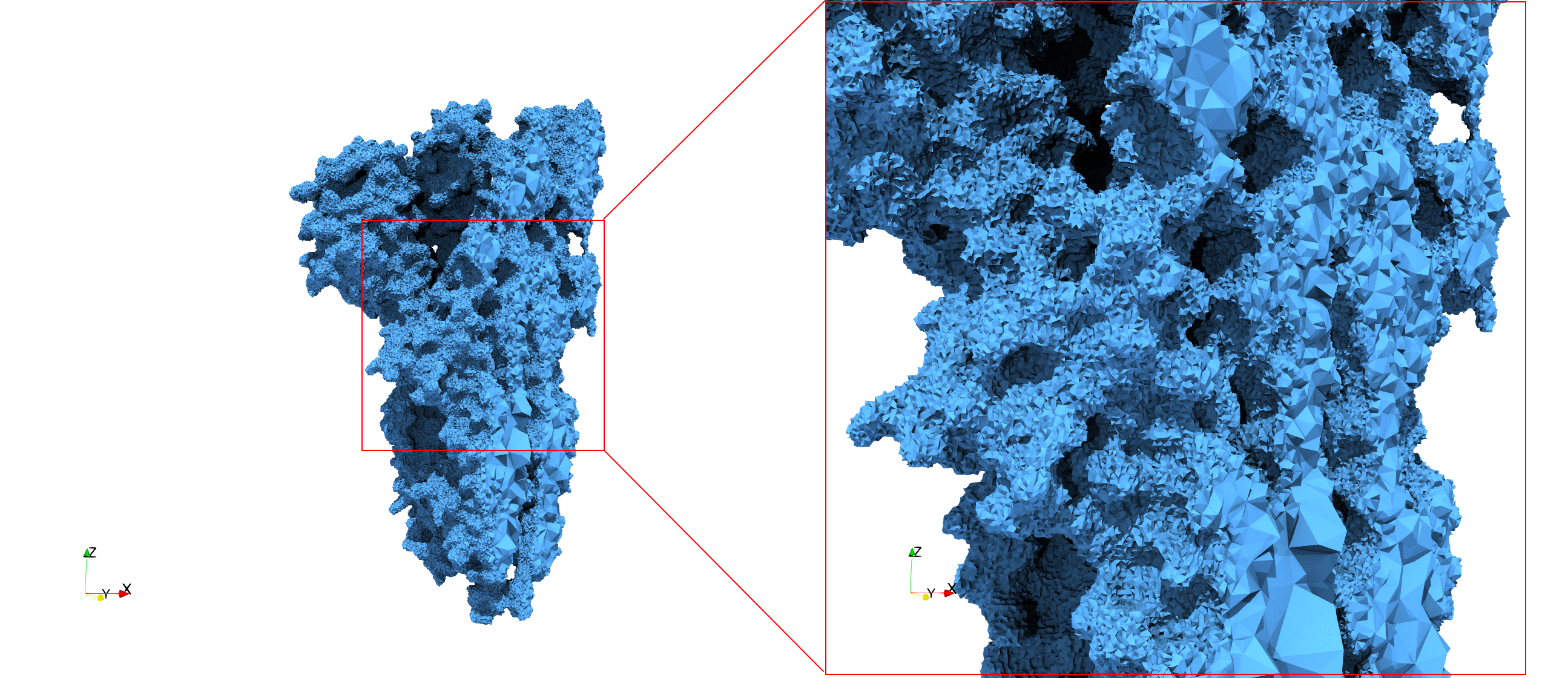
Uniform tetrahedral mesh capable to help identify the internal voids of the <b>COVID-19 NSP15 Endoribonuclease. </b> Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vww 6vww], [https://www.rcsb.org/ Protein Data Bank]. | Uniform tetrahedral mesh capable to help identify the internal voids of the <b>COVID-19 NSP15 Endoribonuclease. </b> Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vww 6vww], [https://www.rcsb.org/ Protein Data Bank]. | ||
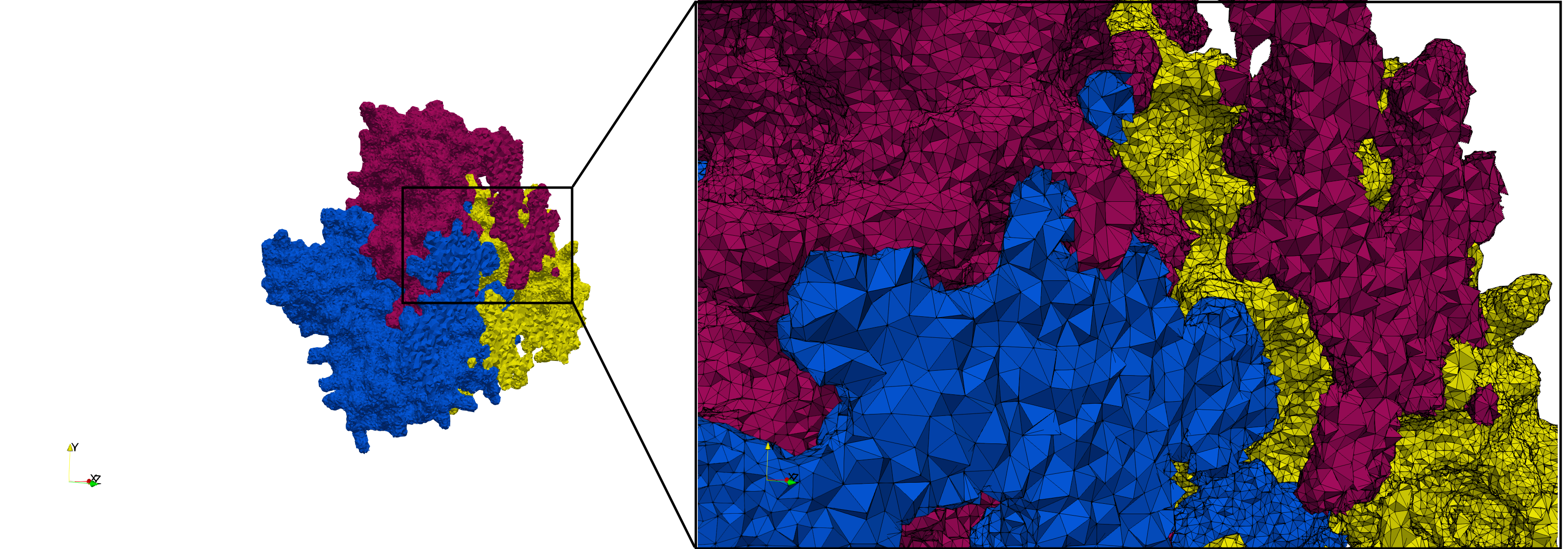
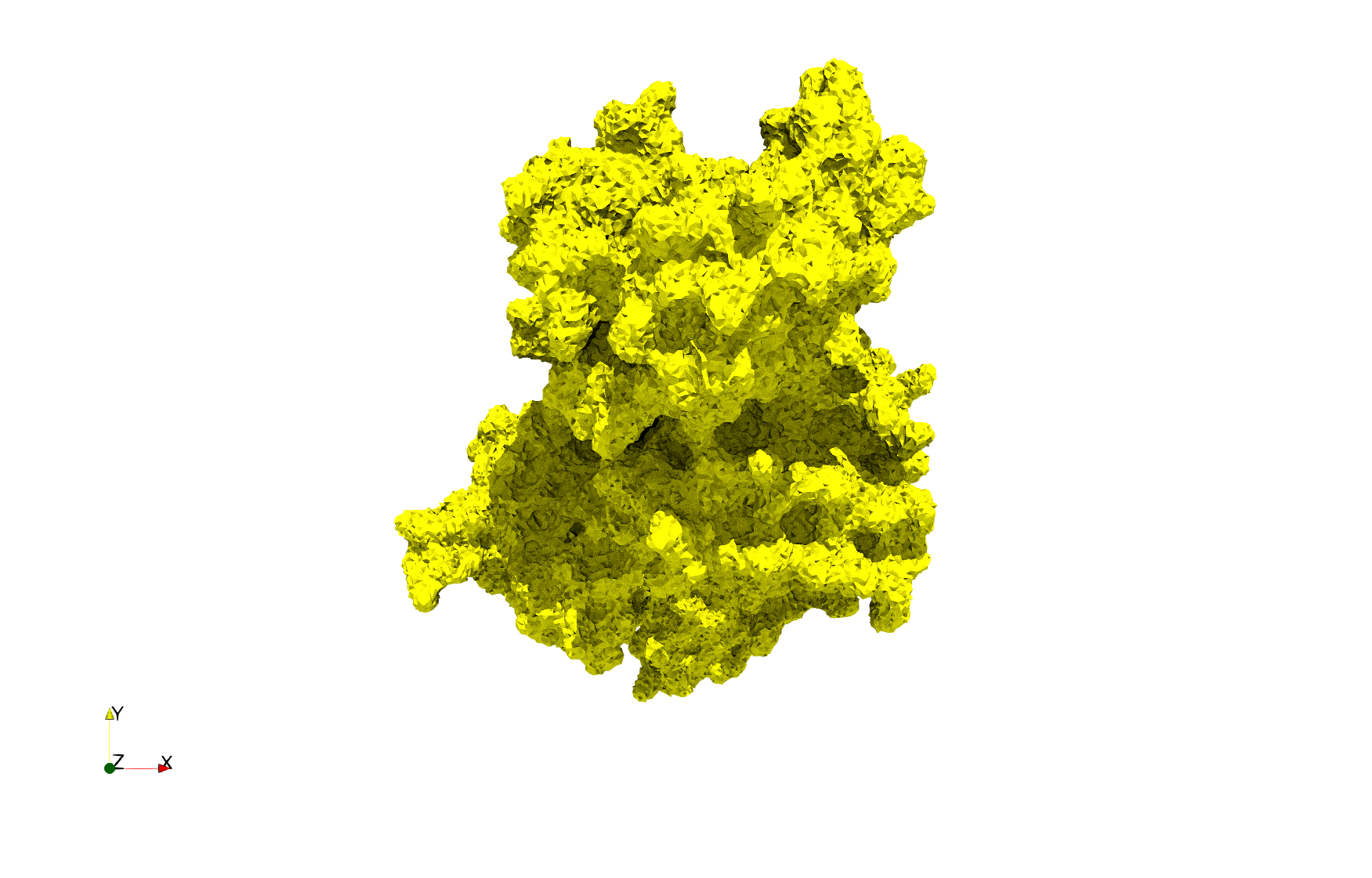
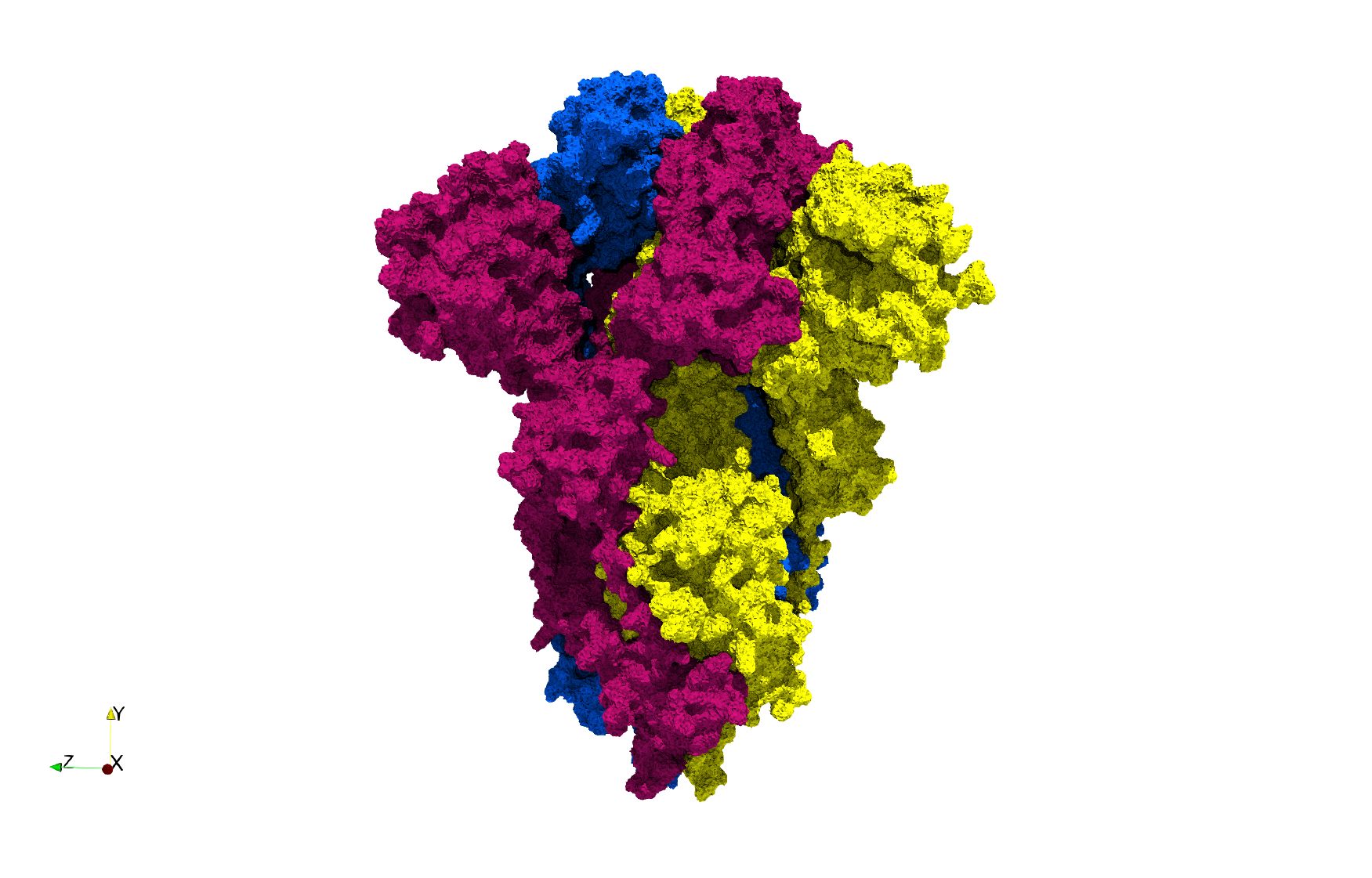
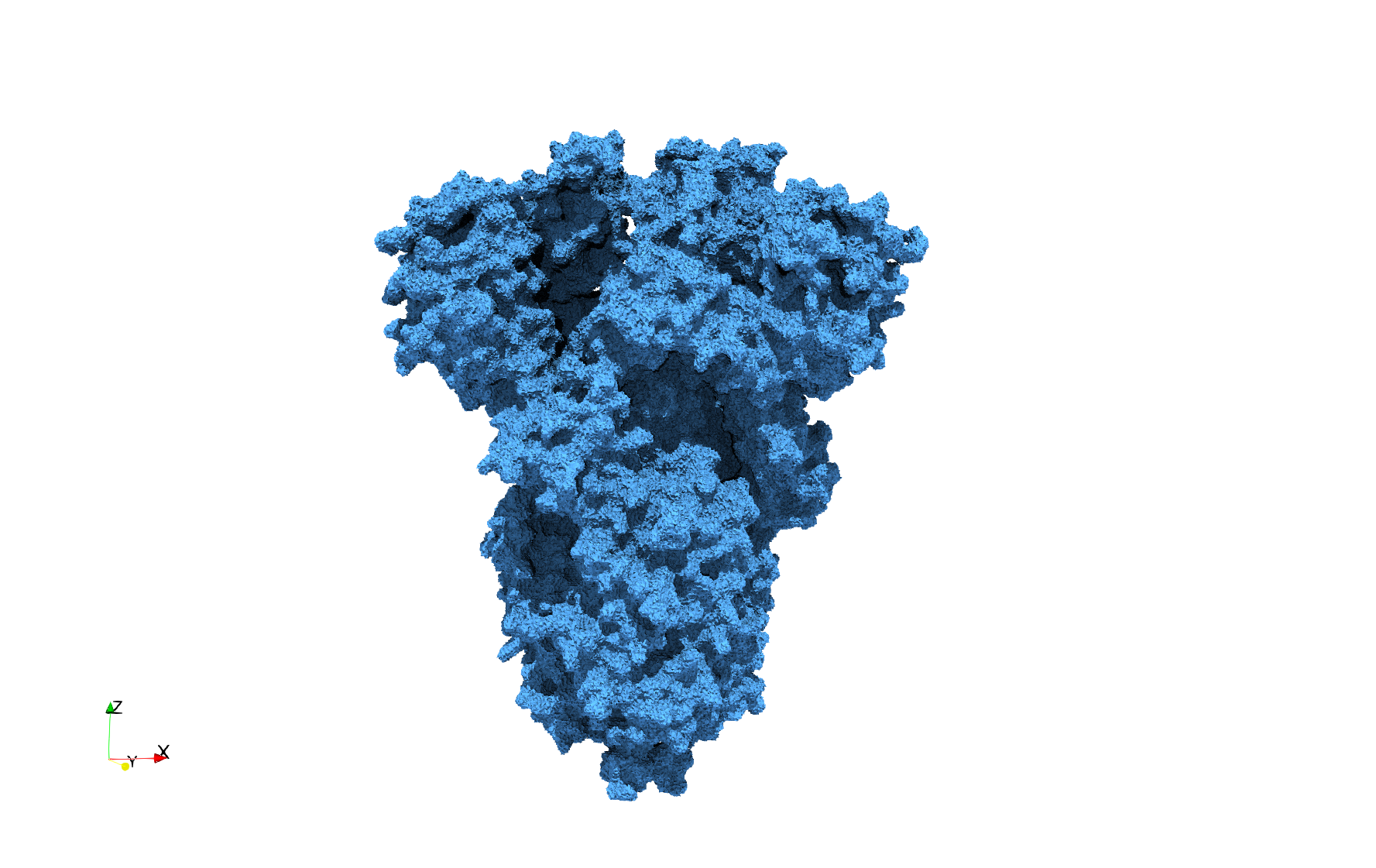
| − | ==COVID-19- | + | ==COVID-19-Spike-Glycoprotein-6vxx== |
| − | * Input image : Dimensions ( | + | * [https://odu.box.com/s/jt2z4vqq3vs8211uuah7m2qfnyk1tjq8 Input Image] |
| − | * Uniform with Delta = 0.5: | + | * Input image : Dimensions (281x380x302) with spacing (0.427871x0.427871x0.427871) |
| + | * Uniform with Delta = 0.5: 3,260,055 tetrahedra | ||
| + | |||
| + | <gallery mode="packed" heights=250px> | ||
| + | File:COVID-19-Spike-Glycoprotein-6vxx,d=5.png | ||
| + | File:COVID-19-Spike-Glycoprotein-6vxx,d=5,zoom.png | ||
| + | </gallery> | ||
<gallery mode="packed" heights=250px> | <gallery mode="packed" heights=250px> | ||
| − | File:COVID-19- | + | File:COVID-19-Spike-Glycoprotein-6vxx,d=5,label=1.png |
| − | File:COVID-19- | + | File:COVID-19-Spike-Glycoprotein-6vxx,d=5,label=2.png |
| − | File:COVID-19- | + | File:COVID-19-Spike-Glycoprotein-6vxx,d=5,label=3.png |
</gallery> | </gallery> | ||
Commands to generate meshes: | Commands to generate meshes: | ||
| − | '''Uniform with Delta = 0.5:''' [https://odu.box.com/s/ | + | '''Uniform with Delta = 0.5:''' [https://odu.box.com/s/0s3zxk3t2hsvmt43l4i9pabm602tiqbk Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19- | + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vxx.nrrd --delta 0.5 --output ./COVID-19-Spike-Glycoprotein-6vxx,d=5.vtk |
</pre> | </pre> | ||
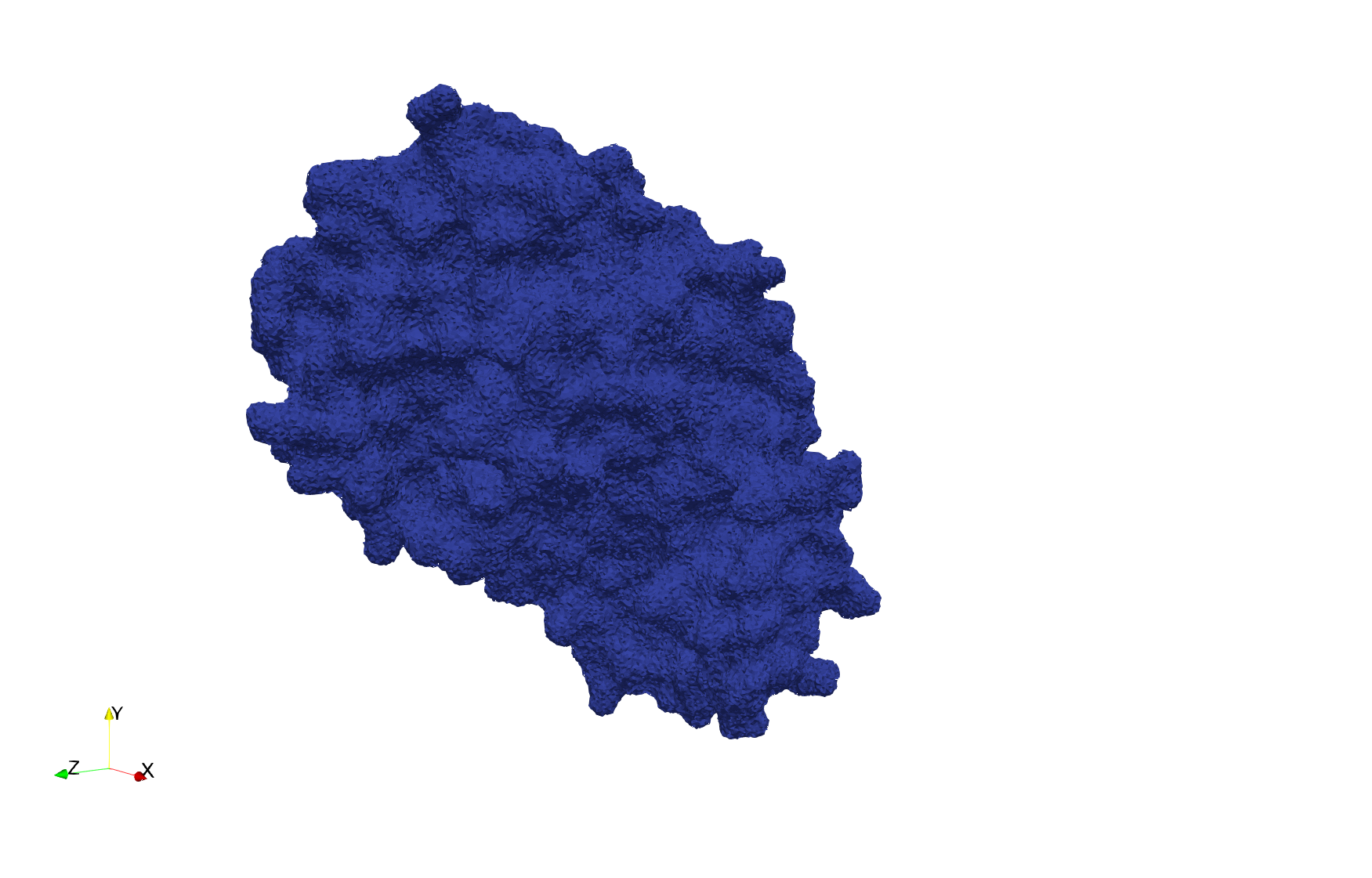
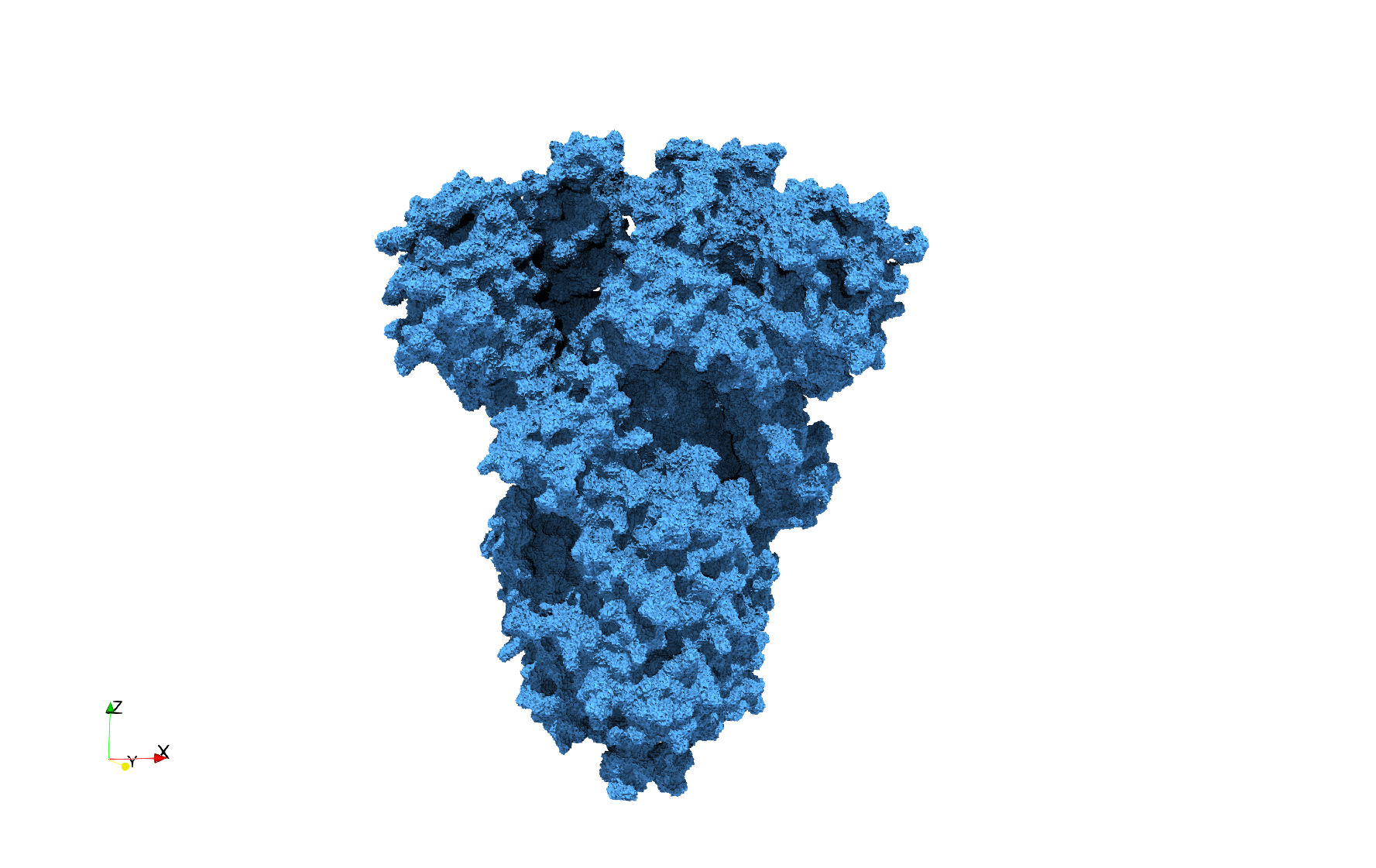
| − | Uniform tetrahedral mesh capable to help identify the internal voids of the <b>COVID-19 | + | Uniform tetrahedral mesh capable to help identify the internal voids of the <b>COVID-19 spike glycoprotein. </b> Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vxx 6vxx], [https://www.rcsb.org/ Protein Data Bank]. |
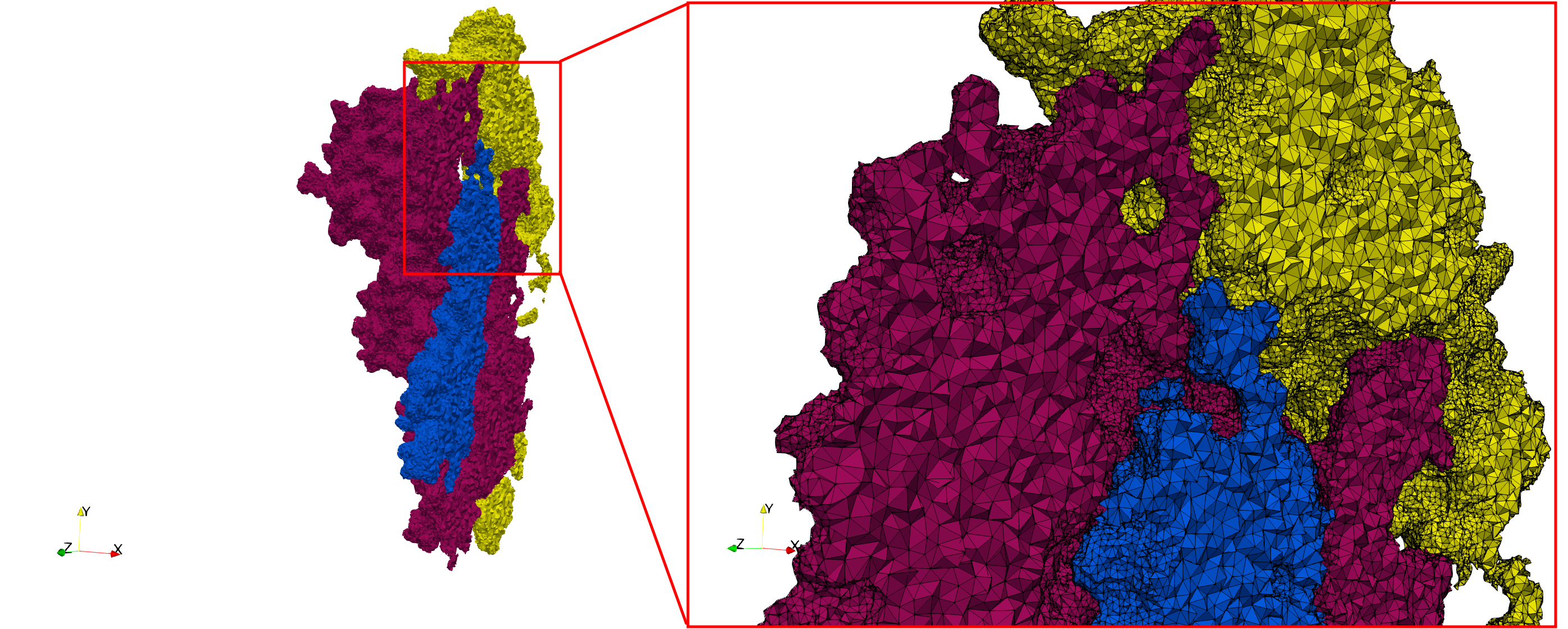
==COVID-19-Spike-Glycoprotein-6vsb== | ==COVID-19-Spike-Glycoprotein-6vsb== | ||
| + | * [https://odu.box.com/s/hebeasclyuekcavgvftb9vbsirrbqj7t Input Image] | ||
* Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042) | * Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042) | ||
* Uniform with Delta = 0.4: 5,731,833 tetrahedra | * Uniform with Delta = 0.4: 5,731,833 tetrahedra | ||
| Line 71: | Line 85: | ||
File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4.png | File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4.png | ||
File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip.png | File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,clip.png | ||
| − | |||
</gallery> | </gallery> | ||
<gallery mode="packed" heights=250px> | <gallery mode="packed" heights=250px> | ||
File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.png | File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.png | ||
File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip.png | File:COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded,clip.png | ||
| − | |||
</gallery> | </gallery> | ||
| Line 92: | Line 104: | ||
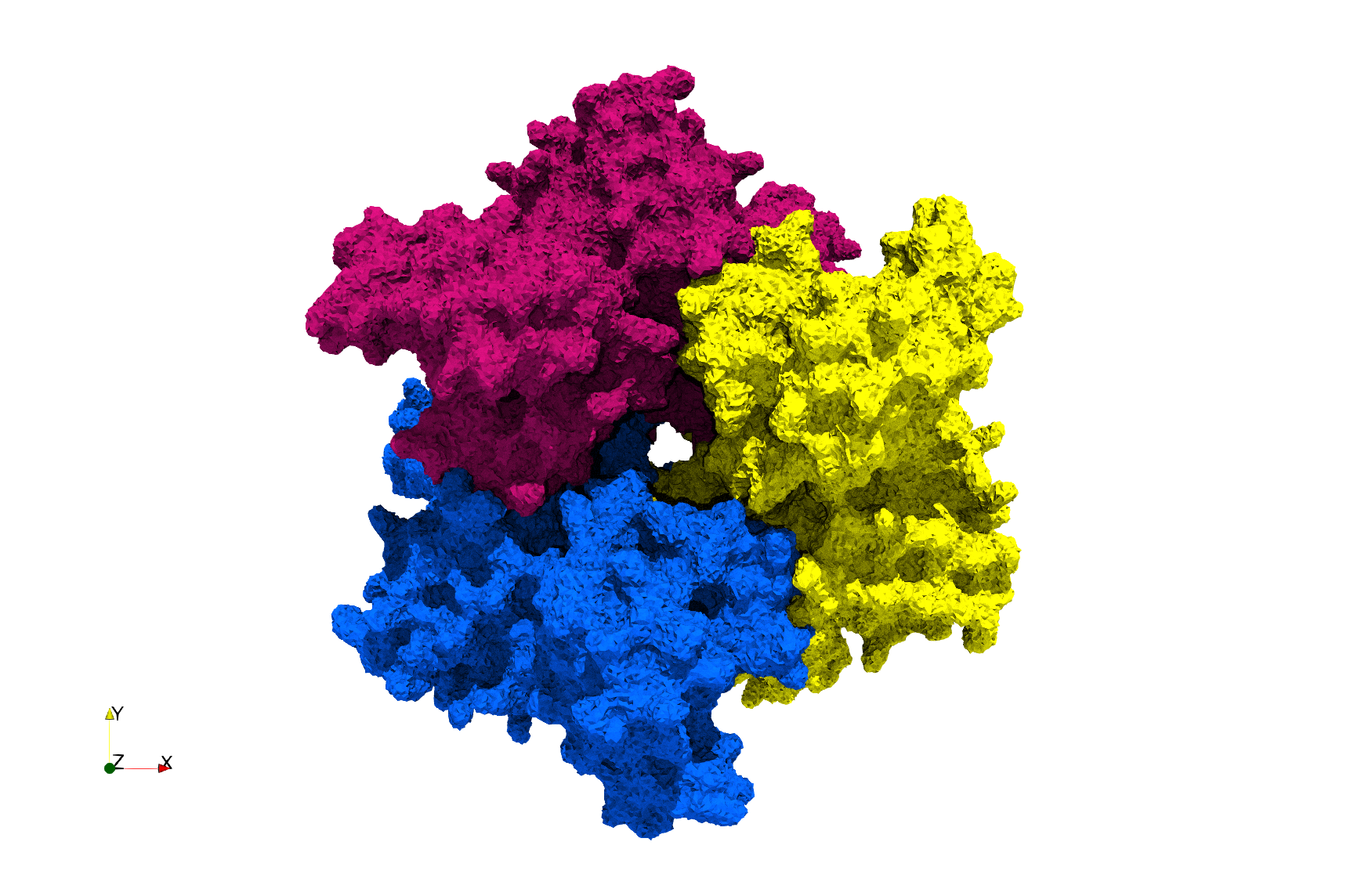
Uniform and graded tetrahedral meshes capable to help identify the internal voids of the <b>COVID-19 spike glycoprotein. </b> The middle and right meshes are slices. Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vxx 6vxx], [https://www.rcsb.org/ Protein Data Bank]. | Uniform and graded tetrahedral meshes capable to help identify the internal voids of the <b>COVID-19 spike glycoprotein. </b> The middle and right meshes are slices. Biological assembly data was retrieved from the molecule [https://www.rcsb.org/structure/6vxx 6vxx], [https://www.rcsb.org/ Protein Data Bank]. | ||
| + | |||
| + | ==Brain-With-Tumor-Case17== | ||
| + | * [https://odu.box.com/s/ynudkxeozt5j82v1qmoovif10c73taw7 Input Image] | ||
| + | * Input image : Dimensions (448x512x176) with spacing (0.488281x0.488281x1) | ||
| + | * Uniform with Delta = default: 222,540 tetrahedra | ||
| + | * Graded with Delta = default: 94,383 tetrahedra | ||
| + | |||
| + | <gallery mode="packed" heights=350px> | ||
| + | File:Brain-With-Tumor-Case17,d=1.76001.png | ||
| + | </gallery> | ||
| + | <gallery mode="packed" heights=350px> | ||
| + | File:Brain-With-Tumor-Case17,d=1.76001,graded.png | ||
| + | </gallery> | ||
| + | |||
| + | Commands to generate meshes: | ||
| + | |||
| + | '''Uniform with Delta = default:''' [https://odu.box.com/s/5dqgw5pykve4xv2ii30xnkjq5s3maxdl Output Mesh] | ||
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --output ./Brain-With-Tumor-Case17,d=1.76001.vtk | ||
| + | </pre> | ||
| + | |||
| + | '''Graded with Delta = default:''' [https://odu.box.com/s/8zlfyepfuzmd8g119eup5n2srxpbqg8s Output Mesh] | ||
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --volume-grading --output ./Brain-With-Tumor-Case17,d=1.76001,graded.vtk | ||
| + | </pre> | ||
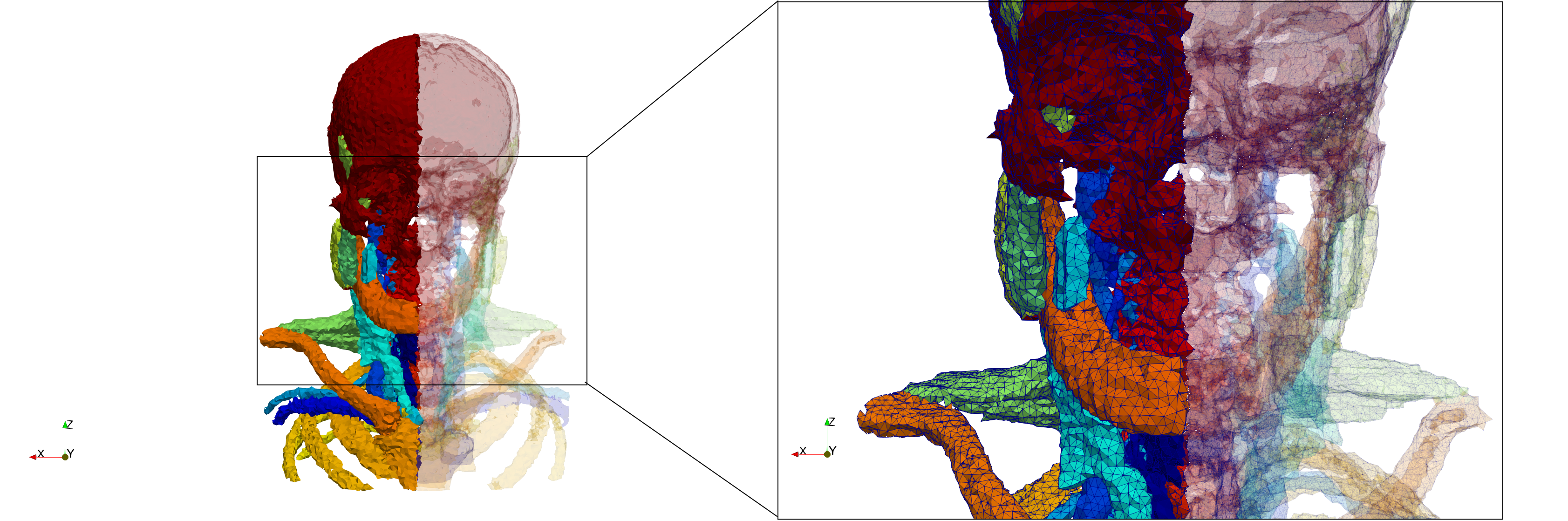
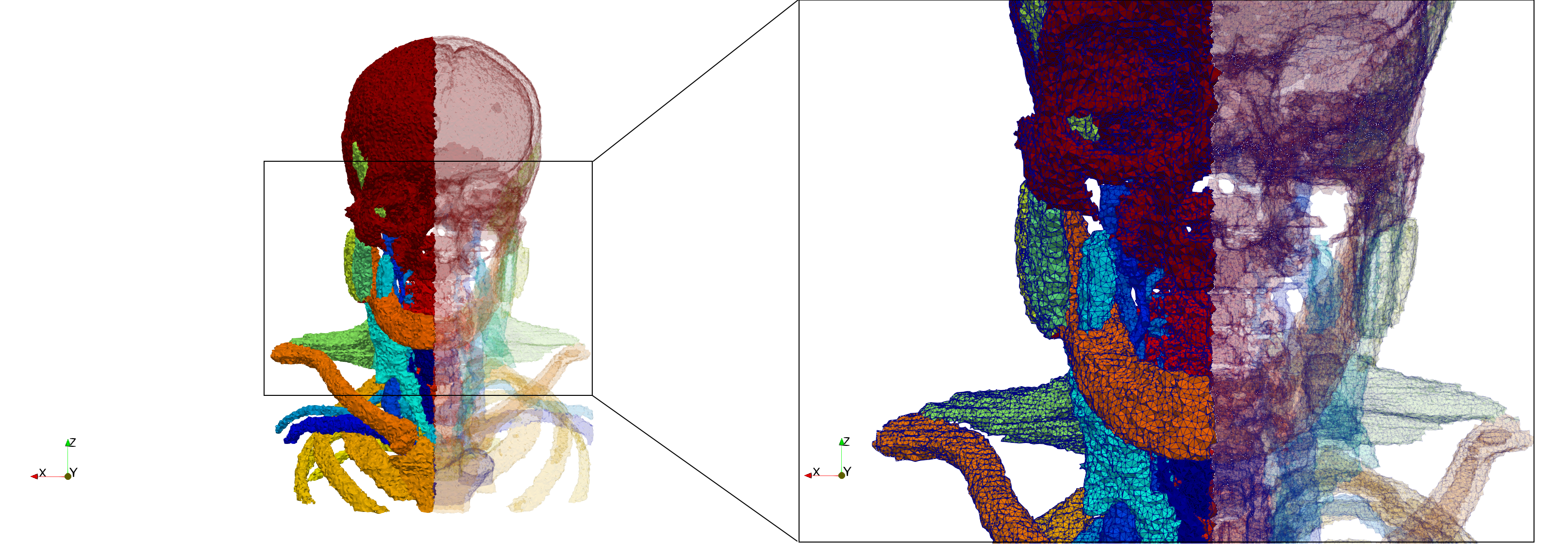
==Head-Neck== | ==Head-Neck== | ||
| + | * [https://odu.box.com/s/zj492g475bo93kyrd1npmisy2gwdqorj Input Image] | ||
* Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002) | * Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002) | ||
* Uniform with Delta = default: 205,510 tetrahedra | * Uniform with Delta = default: 205,510 tetrahedra | ||
| Line 100: | Line 138: | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
File:Head-Neck,d=2.49023.png | File:Head-Neck,d=2.49023.png | ||
| − | |||
</gallery> | </gallery> | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
File:Head-Neck,d=1.5.png | File:Head-Neck,d=1.5.png | ||
| − | |||
</gallery> | </gallery> | ||
| Line 119: | Line 155: | ||
</pre> | </pre> | ||
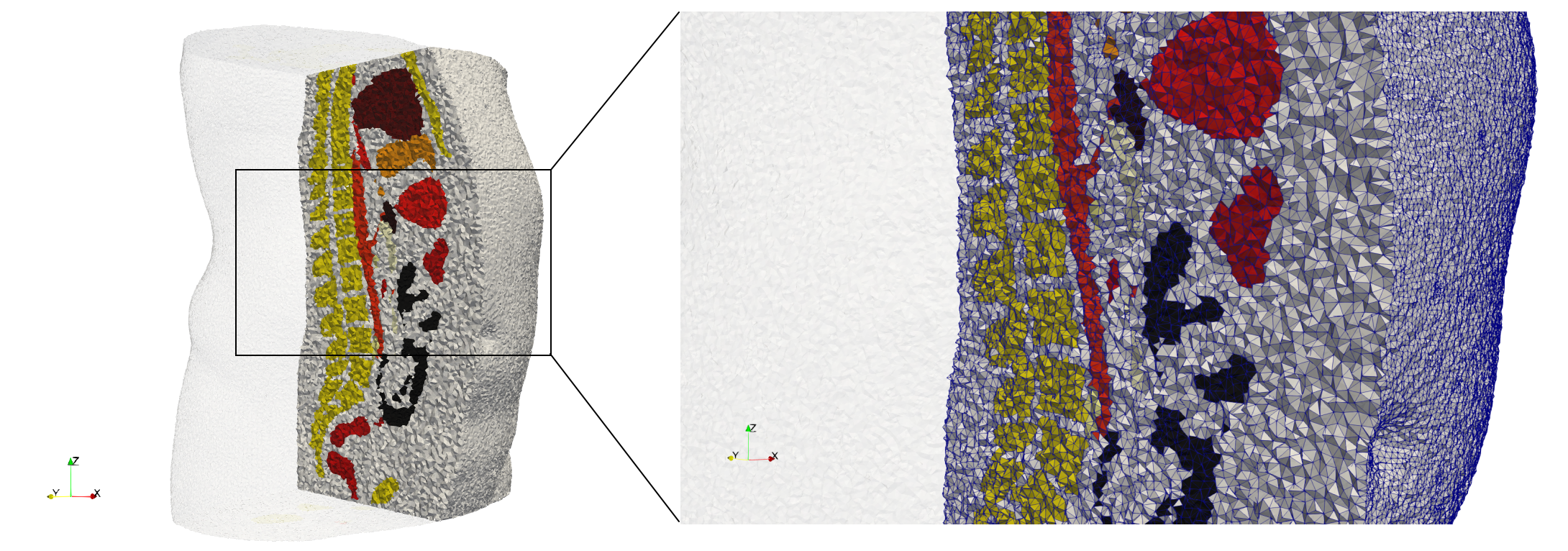
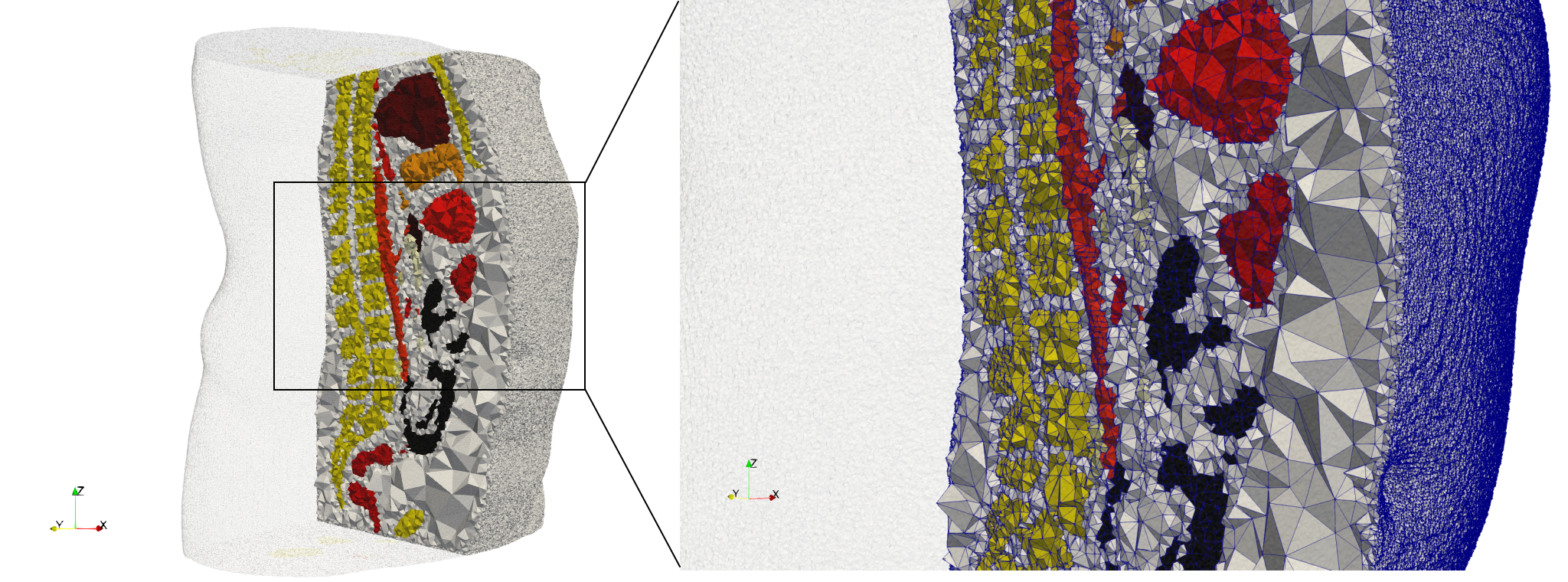
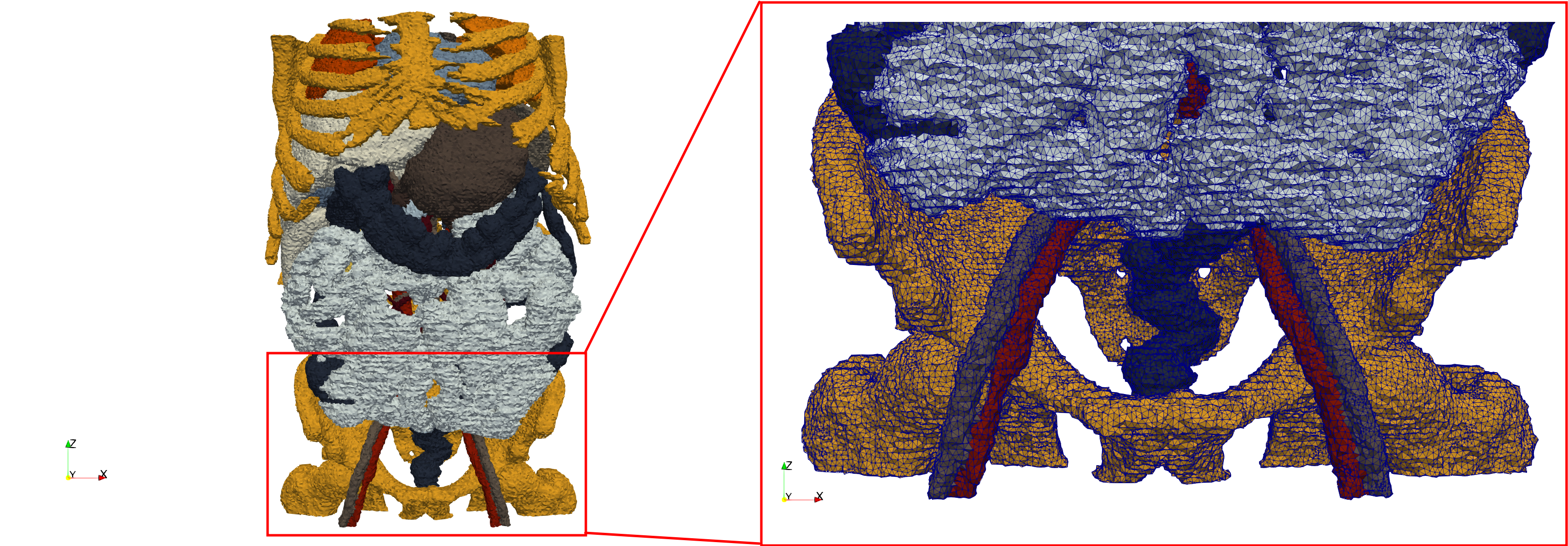
| − | == | + | ==Ircad2== |
| − | * Input image : Dimensions ( | + | * [https://odu.box.com/s/1ef11ry4yik5mny62dz6zyd9obqb540i Input Image] |
| − | * Uniform with Delta = | + | * Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002) |
| − | * Graded with Delta = | + | * Uniform with Delta = 2: 5,031,442 tetrahedra (#elements for each execution might differ due to the usage of more than one thread) |
| + | * Graded with Delta = 1: 6,072,751 tetrahedra (#elements for each execution might differ due to the usage of more than one thread) | ||
| + | * Uniform with Delta = 2 and excluded label 1: 1,941,468 tetrahedra (#elements for each execution might differ due to the usage of more than one thread) | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
| − | File: | + | File:Ircad2,d=2.png |
| − | File: | + | </gallery> |
| + | <gallery mode="packed" heights=350px> | ||
| + | File:Ircad2,d=1,graded.png | ||
</gallery> | </gallery> | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
| − | File: | + | File:Ircad2,d=2,el=1.png |
| − | |||
</gallery> | </gallery> | ||
Commands to generate meshes: | Commands to generate meshes: | ||
| − | '''Uniform with Delta = | + | '''Uniform with Delta = 2:''' [https://odu.box.com/s/nfqgcr1j1j6wdnydb9sug8652y9falvc Output Mesh] |
| + | <pre> | ||
| + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 2 --threads 6 --output ./Ircad2,d=2.vtk | ||
| + | </pre> | ||
| + | |||
| + | '''Graded with Delta = 1:''' [https://odu.box.com/s/ofnxslfgijzbqbwqqlvcetm783kqv9uo Output Mesh] | ||
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/ | + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 6 --volume-grading --output ./Ircad2,d=1,graded.vtk |
</pre> | </pre> | ||
| − | ''' | + | '''Uniform with Delta = 2 and excluded label 1:''' [https://odu.box.com/s/5jaukd6317jdmi8li479h8emknuw0rny Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/ | + | docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --exclude-labels 1 --delta 2 --threads 6 --output ./Ircad2,d=2,el=1.vtk |
</pre> | </pre> | ||
==Knee-Char== | ==Knee-Char== | ||
| + | * [https://odu.box.com/s/jv3rqazsth3ktd8ipsstzqjl6ryz8naf Input Image] | ||
* Input image : Dimensions (512x512x119) with spacing (0.27734x0.27734x1) | * Input image : Dimensions (512x512x119) with spacing (0.27734x0.27734x1) | ||
* Uniform with Delta = default : 386,869 tetrahedra | * Uniform with Delta = default : 386,869 tetrahedra | ||
| Line 152: | Line 197: | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
File:Knee-Char,d=1.19.png | File:Knee-Char,d=1.19.png | ||
| − | |||
</gallery> | </gallery> | ||
<gallery mode="packed" heights=350px> | <gallery mode="packed" heights=350px> | ||
File:Knee-Char,d=1.19,graded.png | File:Knee-Char,d=1.19,graded.png | ||
| − | |||
</gallery> | </gallery> | ||
| Line 171: | Line 214: | ||
</pre> | </pre> | ||
| − | == | + | =2D Example Meshes= |
| − | * Input image : Dimensions ( | + | The directory containing the 2D input data is located in the 2D folder of [https://odu.box.com/s/olefferrnksu2nmerbfvbvsz2u4abbdw Medical_Imaging_Data]. |
| − | * Uniform with | + | |
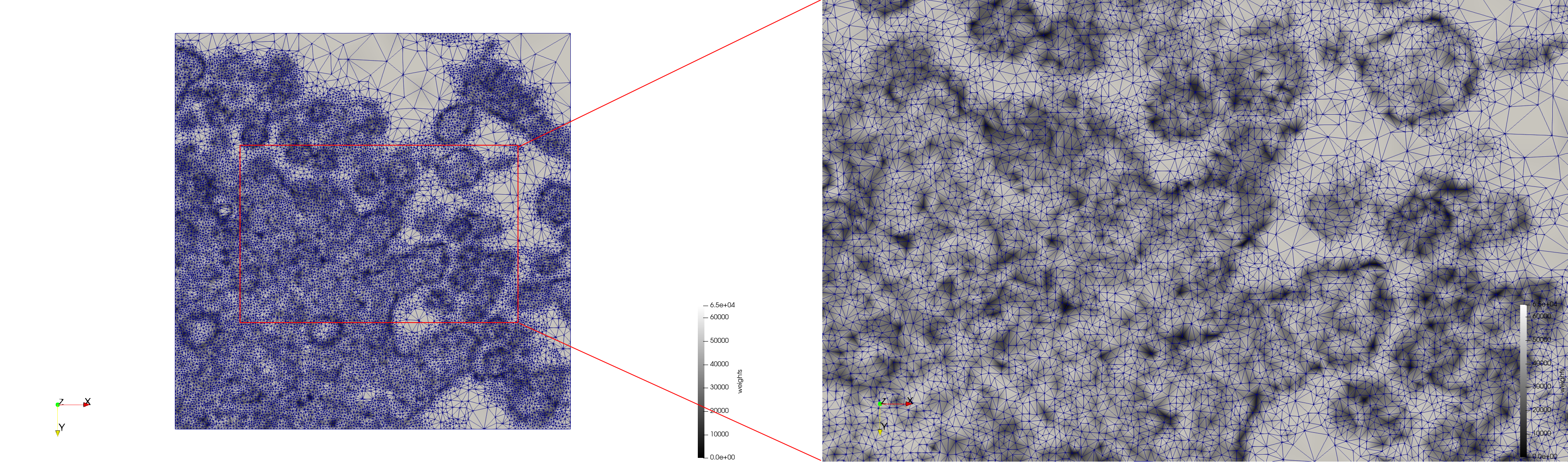
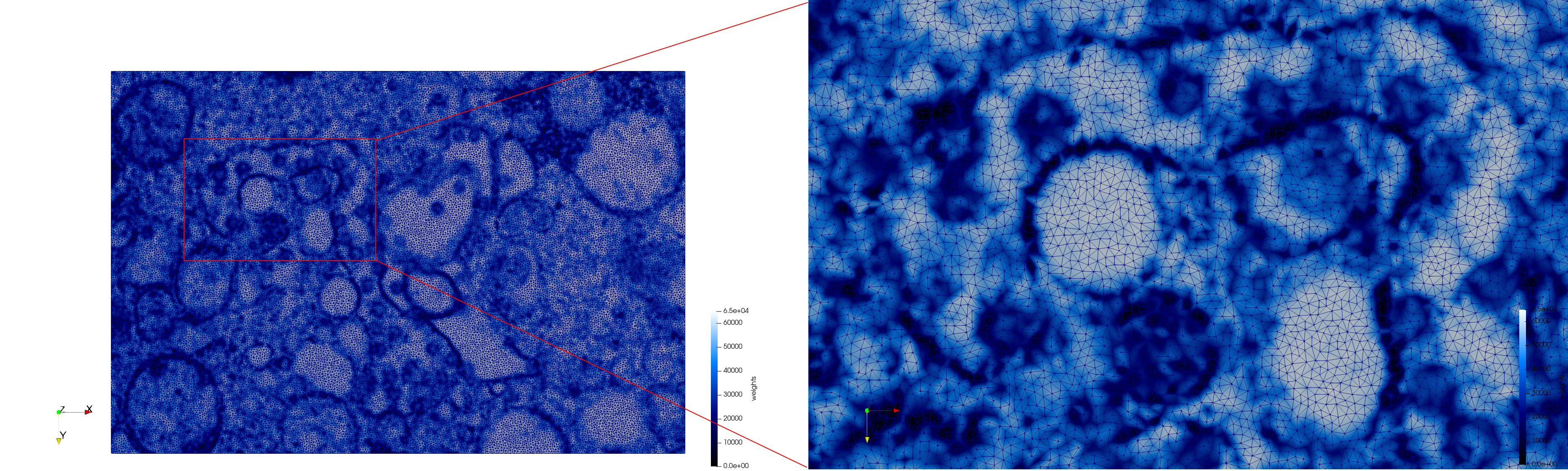
| − | * | + | ==COVID-19-23311== |
| + | * [https://odu.box.com/s/musv3zdpx6a76p282t2xn905rf0xlwty Input Image] | ||
| + | * Input image : Dimensions (2,460x2,460) with spacing (1x1) | ||
| + | * Uniform with Min-Edge = 20: 47,028 triangles | ||
| + | * Adaptive with Min-Edge = 20: 34,080 tetrahedra | ||
| − | File: | + | <gallery mode="packed" heights=250px> |
| − | File: | + | File:COVID-19-23311,uniform,e=20,suface.png |
| + | File:COVID-19-23311,uniform,e=20,triangulation.png | ||
| + | </gallery> | ||
| + | <gallery mode="packed" heights=250px> | ||
| + | File:COVID-19-23311,w=0.1,e=20,suface.png | ||
| + | File:COVID-19-23311,w=0.1,e=20,triangulation.png | ||
| + | </gallery> | ||
Commands to generate meshes: | Commands to generate meshes: | ||
| − | '''Uniform with | + | '''Uniform with Min-Edge = 20:''' [https://odu.box.com/s/dkv6271ky0cxvk4lio2bvw6jv9zw5iaf Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m | + | docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --uniform --min-edge 20 --output ./COVID-19-23311,uniform,e=20.vtk |
</pre> | </pre> | ||
| − | ''' | + | '''Adaptive with Min-Edge = 20:''' [https://odu.box.com/s/jeqmz2c8dsssl8zujkywl2v3bjaby8gc Output Mesh] |
<pre> | <pre> | ||
| − | docker run -v $(pwd):/data/ crtc_i2m | + | docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --min-edge 20 --output ./COVID-19-23311,w=0.1,e=20.vtk |
</pre> | </pre> | ||
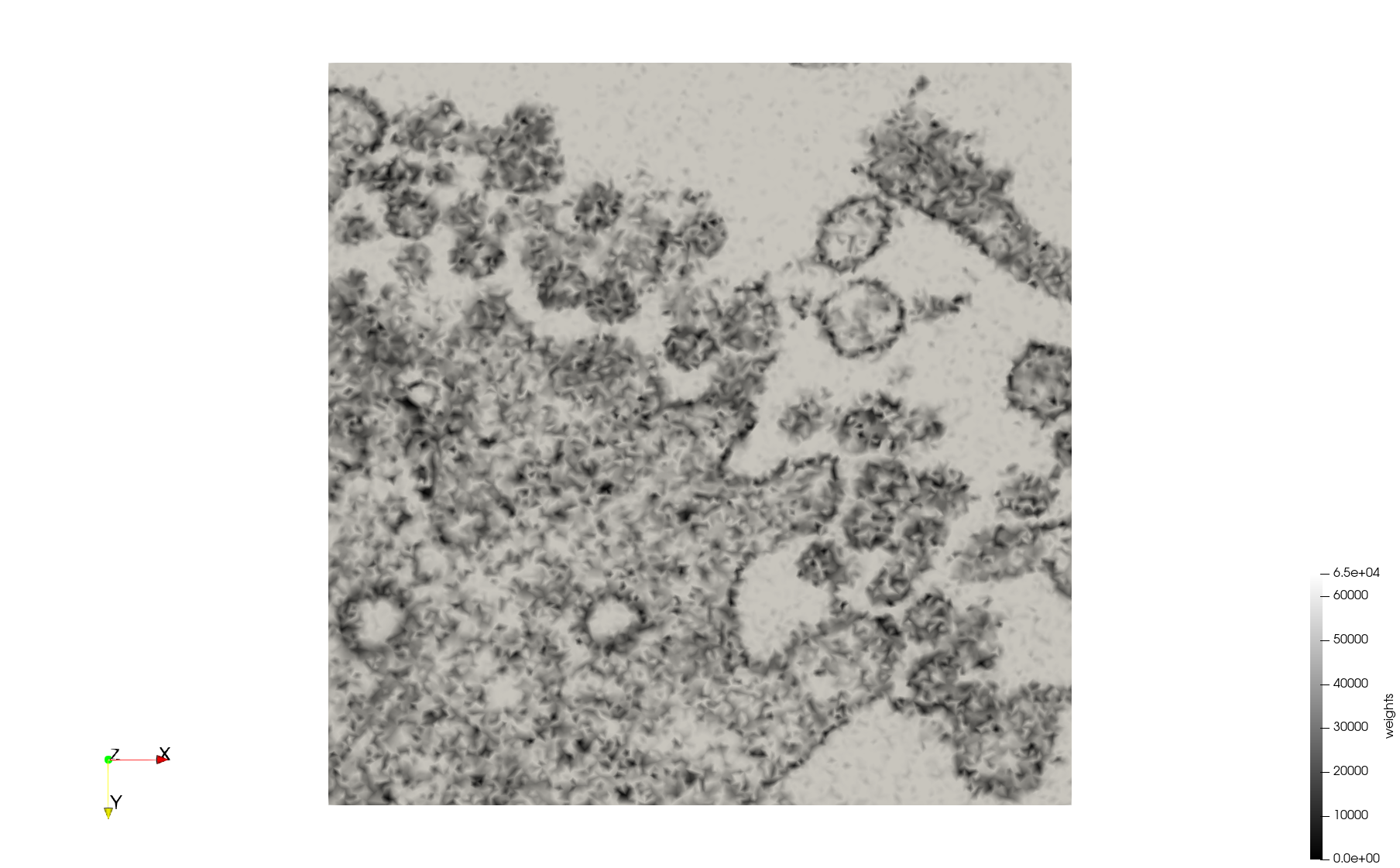
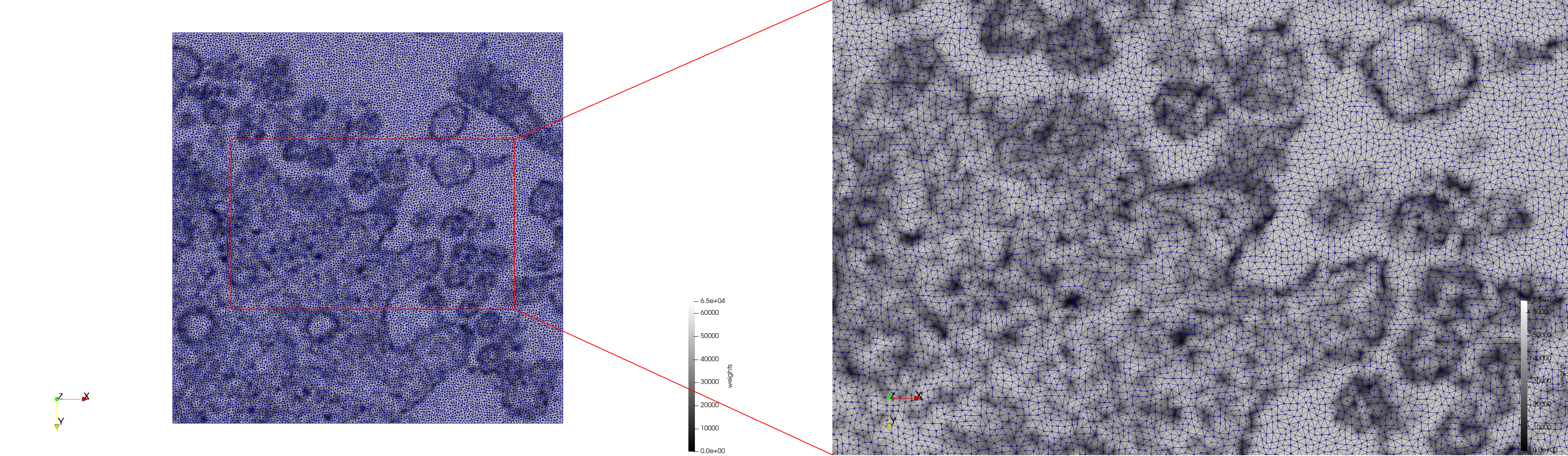
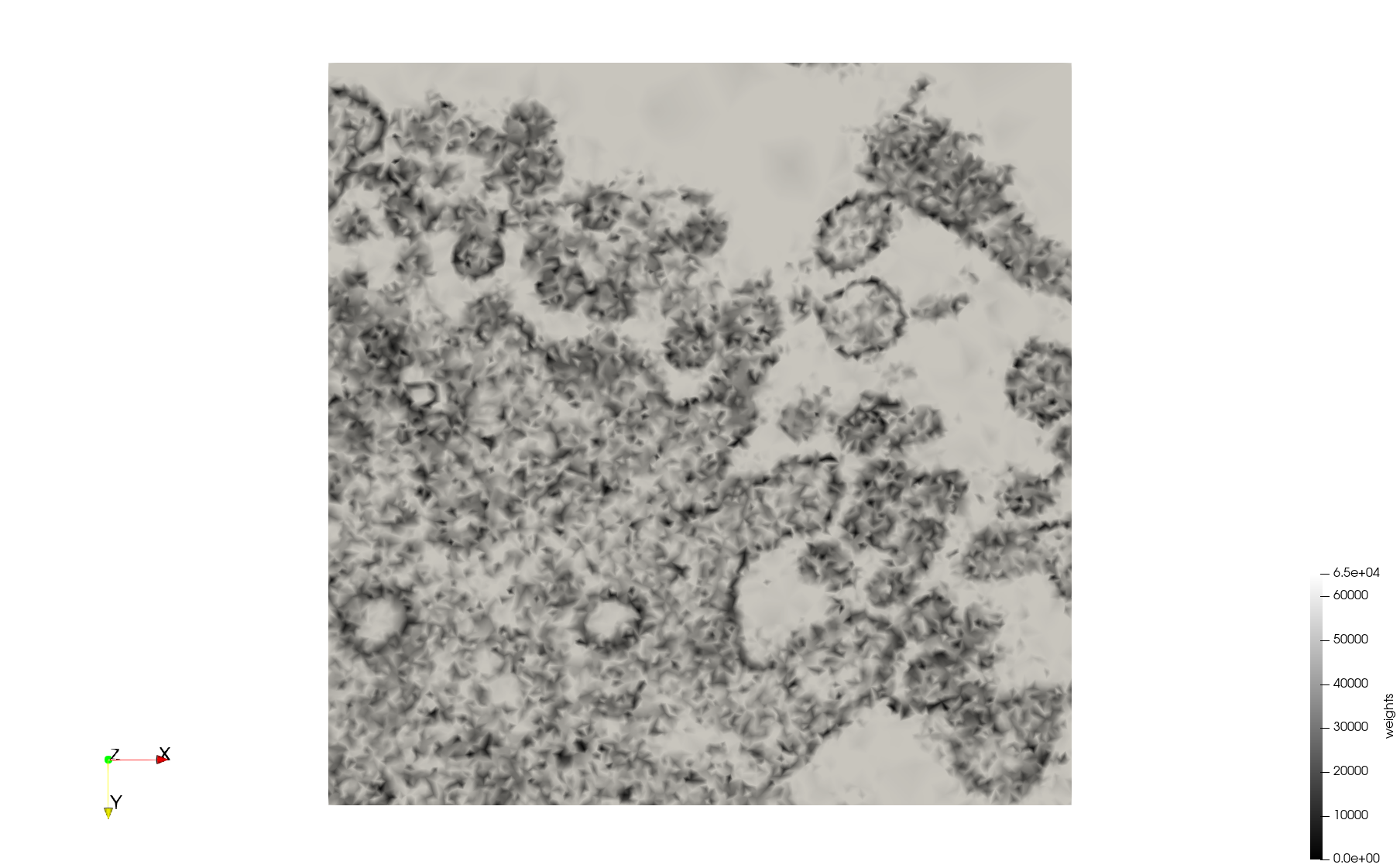
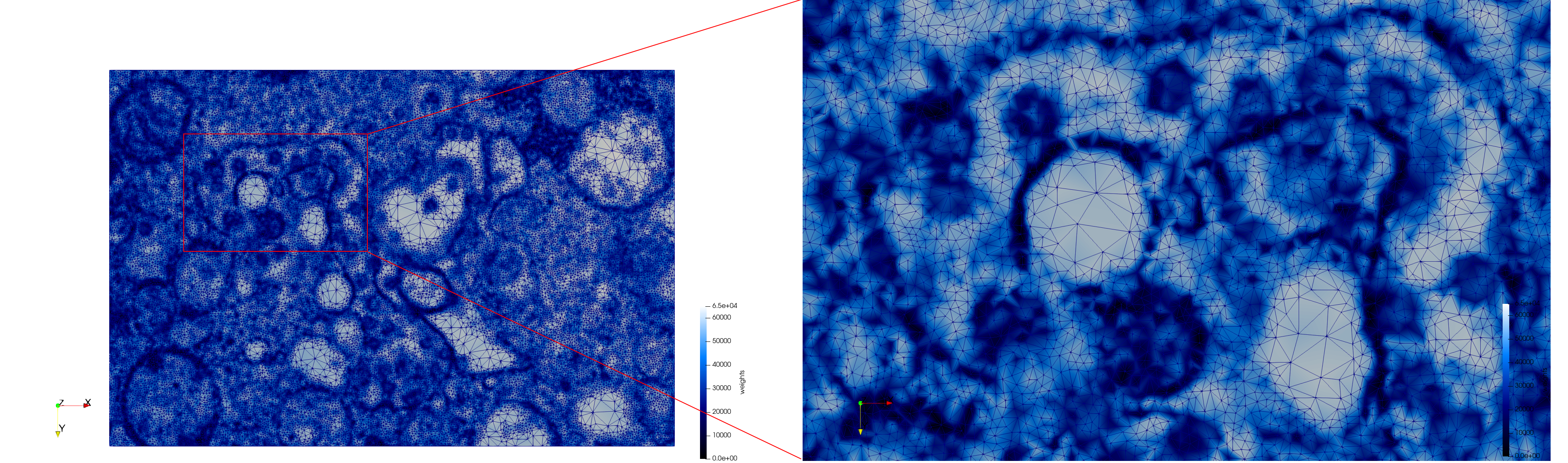
| − | + | Meshes generated based on the image [https://phil.cdc.gov//PHIL_Images/23311/23311.tif 23311] retrieved from the [https://www.cdc.gov/media/subtopic/images.htm Centers for Disease Control and Prevention]. For the uniform mesh, the edge-size corresponds to 20 pixels. The adaptive was created by controlling the size of the elements based on the difference in the intensity of the pixels. | |
| − | |||
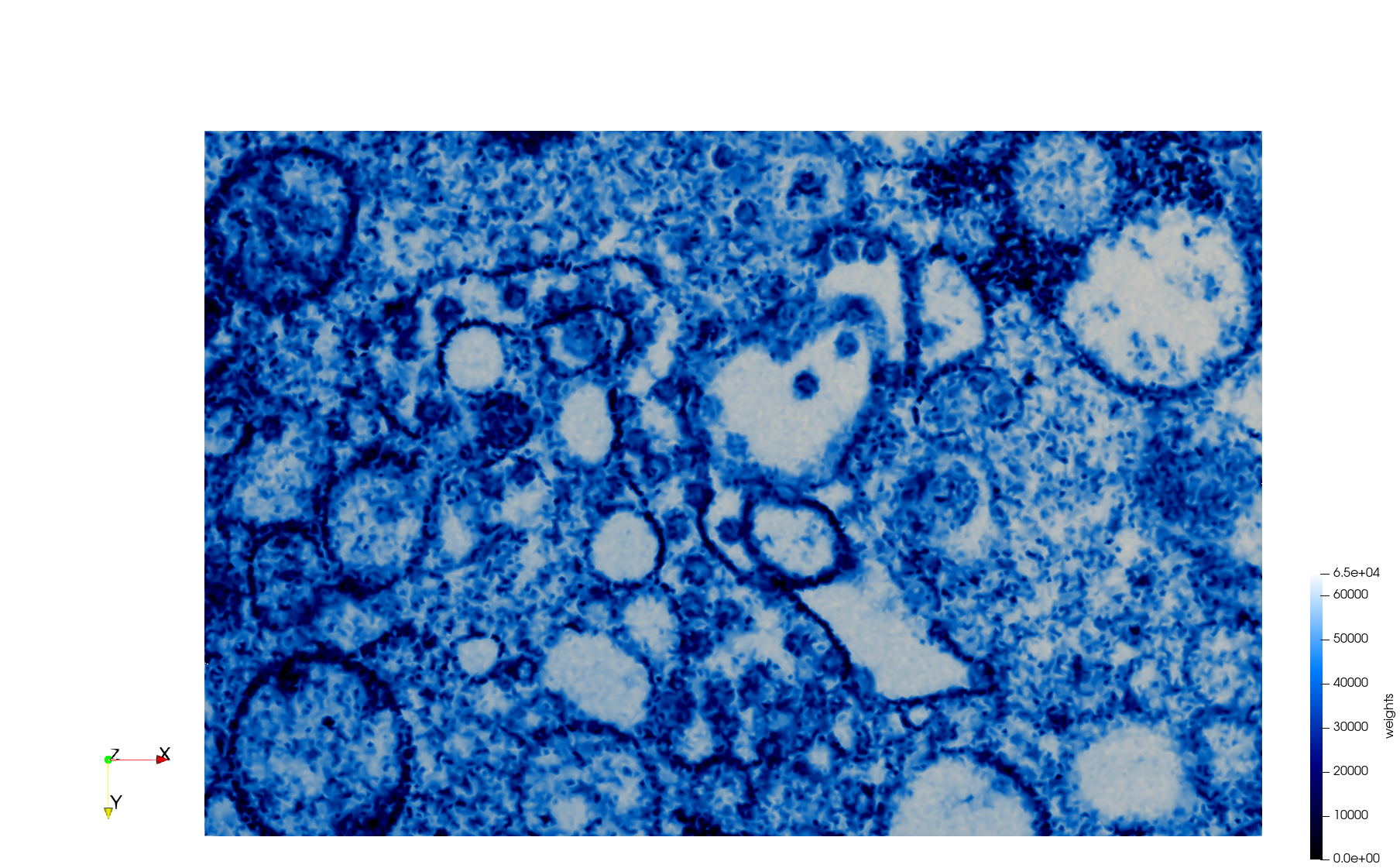
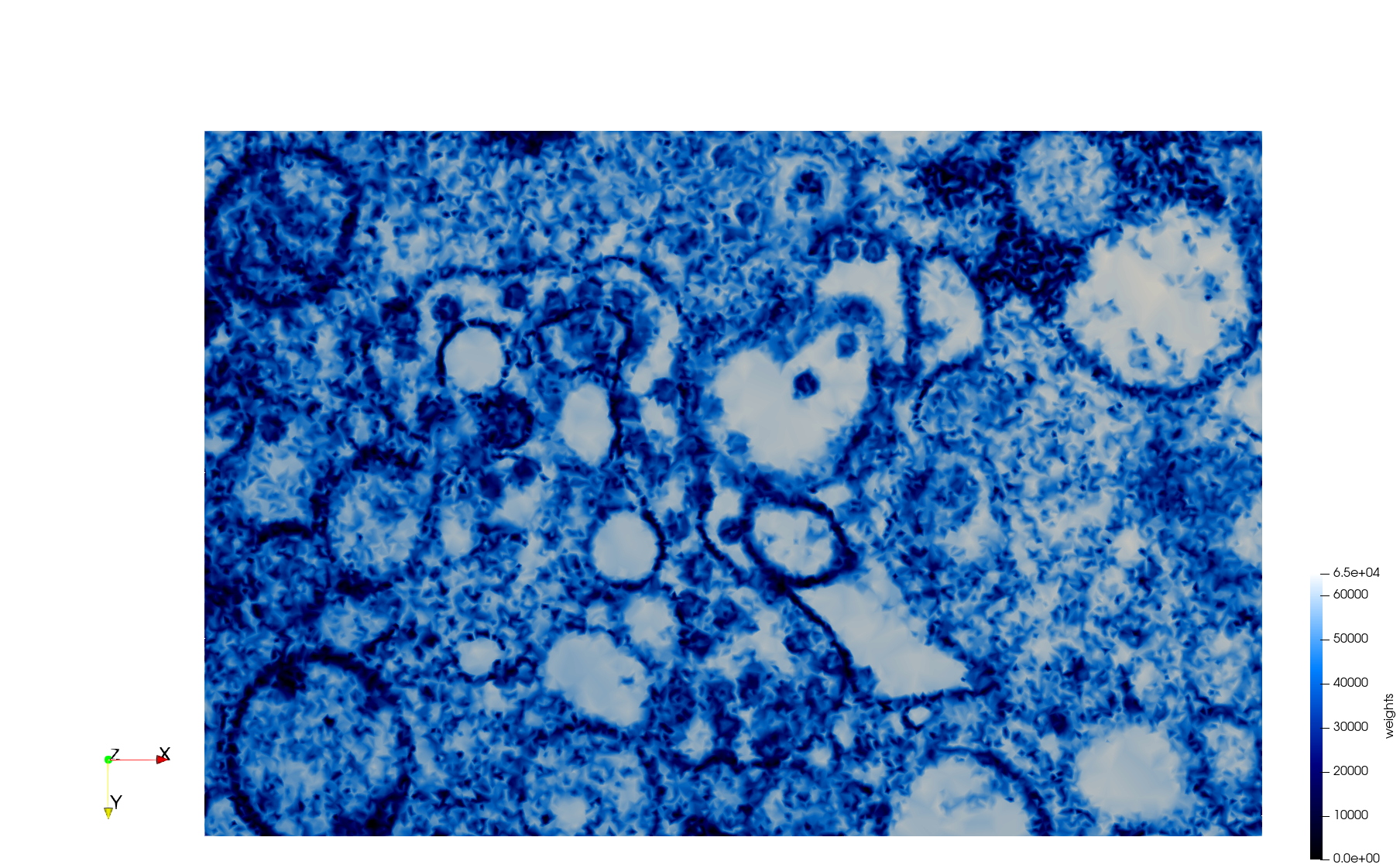
==COVID-19-23354== | ==COVID-19-23354== | ||
| + | * [https://odu.box.com/s/vwse25rtxemv27sf8vkzofupc4l5mnc7 Input Image] | ||
* Input image : Dimensions (3,000x2,000) with spacing (1x1) | * Input image : Dimensions (3,000x2,000) with spacing (1x1) | ||
* Uniform with Min-Edge = 15: 82,981 triangles | * Uniform with Min-Edge = 15: 82,981 triangles | ||
| Line 220: | Line 273: | ||
</pre> | </pre> | ||
| − | + | Meshes generated based on the image [https://phil.cdc.gov//PHIL_Images/23354/23354.tif 23354] retrieved from the [https://www.cdc.gov/media/subtopic/images.htm Centers for Disease Control and Prevention]. For the uniform mesh, the edge-size corresponds to 15 pixels. The adaptive was created by controlling the size of the elements based on the difference in the intensity of the pixels. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 16:01, 20 April 2020
Contents
3D Example Meshes
The directory containing the 3D input data is located in the 3D folder of Medical_Imaging_Data.
COVID-19-Main-Protease-6y2e
- Input Image
- Input image : Dimensions (284x303x344) with spacing (0.2226907x0.2226907x0.2226907)
- Uniform with Delta = 0.3: 2,509,202 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.3: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Main-Protease-6y2e.nrrd --delta 0.3 --output ./COVID-19-Main-Protease-6y2e,d=0.3.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 Main Protease. Biological assembly data was retrieved from the molecule 6y2e, Protein Data Bank.
COVID-19-NSP-15-Endoribonuclease-6vww
- [Input Image]
- Input image : Dimensions (303x297x337) with spacing (0.3445713x0.3445713x0.3445713)
- Uniform with Delta = 0.5: 2,310,215 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-NSP-15-Endoribonuclease-6vww.nrrd --delta 0.5 --output ./COVID-19-NSP-15-Endoribonuclease-6vww,d=0.5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 NSP15 Endoribonuclease. Biological assembly data was retrieved from the molecule 6vww, Protein Data Bank.
COVID-19-Spike-Glycoprotein-6vxx
- Input Image
- Input image : Dimensions (281x380x302) with spacing (0.427871x0.427871x0.427871)
- Uniform with Delta = 0.5: 3,260,055 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vxx.nrrd --delta 0.5 --output ./COVID-19-Spike-Glycoprotein-6vxx,d=5.vtk
Uniform tetrahedral mesh capable to help identify the internal voids of the COVID-19 spike glycoprotein. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
COVID-19-Spike-Glycoprotein-6vsb
- Input Image
- Input image : Dimensions (220x223x314) with spacing (0.551042x0.551042x0.551042)
- Uniform with Delta = 0.4: 5,731,833 tetrahedra
- Graded with Delta = 0.4: 3,794,223 tetrahedra
Commands to generate meshes:
Uniform with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4.vtk
Graded with Delta = 0.4: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/COVID-19-Spike-Glycoprotein-6vsb.nrrd --delta 0.4 --volume-grading --output ./COVID-19-Spike-Glycoprotein-6vsb,d=0.4,graded.vtk
Uniform and graded tetrahedral meshes capable to help identify the internal voids of the COVID-19 spike glycoprotein. The middle and right meshes are slices. Biological assembly data was retrieved from the molecule 6vxx, Protein Data Bank.
Brain-With-Tumor-Case17
- Input Image
- Input image : Dimensions (448x512x176) with spacing (0.488281x0.488281x1)
- Uniform with Delta = default: 222,540 tetrahedra
- Graded with Delta = default: 94,383 tetrahedra
Commands to generate meshes:
Uniform with Delta = default: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --output ./Brain-With-Tumor-Case17,d=1.76001.vtk
Graded with Delta = default: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Brain-With-Tumor-Case17.nii --volume-grading --output ./Brain-With-Tumor-Case17,d=1.76001,graded.vtk
Head-Neck
- Input Image
- Input image : Dimensions (255x255x229) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = default: 205,510 tetrahedra
- Uniform with Delta = 1.5: 767,393 tetrahedra
Commands to generate meshes:
Uniform with Delta = default: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --output ./Head-Neck,d=2.49023.vtk
Uniform with Delta = 1.5: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Head-Neck.mha --delta 1.5 --output ./Head-Neck,d=1.5.vtk
Ircad2
- Input Image
- Input image : Dimensions (512x512x219) with spacing (0.976562x0.976562x1.40002)
- Uniform with Delta = 2: 5,031,442 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
- Graded with Delta = 1: 6,072,751 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
- Uniform with Delta = 2 and excluded label 1: 1,941,468 tetrahedra (#elements for each execution might differ due to the usage of more than one thread)
Commands to generate meshes:
Uniform with Delta = 2: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 2 --threads 6 --output ./Ircad2,d=2.vtk
Graded with Delta = 1: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --delta 1 --threads 6 --volume-grading --output ./Ircad2,d=1,graded.vtk
Uniform with Delta = 2 and excluded label 1: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Ircad2.nrrd --exclude-labels 1 --delta 2 --threads 6 --output ./Ircad2,d=2,el=1.vtk
Knee-Char
- Input Image
- Input image : Dimensions (512x512x119) with spacing (0.27734x0.27734x1)
- Uniform with Delta = default : 386,869 tetrahedra
- Graded with Delta = default : 274,309 tetrahedra
Commands to generate meshes:
Uniform with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Knee-Char.mha --output ./Knee-Char,d=1.19.vtk
Graded with Delta = default : Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate3d --input ./Medical_Imaging_Data/3D/Knee-Char.mha --volume-grading --output ./Knee-Char,d=1.19,graded.vtk
2D Example Meshes
The directory containing the 2D input data is located in the 2D folder of Medical_Imaging_Data.
COVID-19-23311
- Input Image
- Input image : Dimensions (2,460x2,460) with spacing (1x1)
- Uniform with Min-Edge = 20: 47,028 triangles
- Adaptive with Min-Edge = 20: 34,080 tetrahedra
Commands to generate meshes:
Uniform with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --uniform --min-edge 20 --output ./COVID-19-23311,uniform,e=20.vtk
Adaptive with Min-Edge = 20: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23311.tif --min-edge 20 --output ./COVID-19-23311,w=0.1,e=20.vtk
Meshes generated based on the image 23311 retrieved from the Centers for Disease Control and Prevention. For the uniform mesh, the edge-size corresponds to 20 pixels. The adaptive was created by controlling the size of the elements based on the difference in the intensity of the pixels.
COVID-19-23354
- Input Image
- Input image : Dimensions (3,000x2,000) with spacing (1x1)
- Uniform with Min-Edge = 15: 82,981 triangles
- Adaptive with Min-Edge = 15: 67,920 triangles
Commands to generate meshes:
Uniform with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --uniform --min-edge 15 --output ./COVID-19-23354,uniform,e=15.vtk
Adaptive with Min-Edge = 15: Output Mesh
docker run -v $(pwd):/data/ crtc_i2m tessellate2d --input ./Medical_Imaging_Data/2D/COVID-19-23354.tif --min-edge 15 --output ./COVID-19-23354,w=0.1,e=15.vtk
Meshes generated based on the image 23354 retrieved from the Centers for Disease Control and Prevention. For the uniform mesh, the edge-size corresponds to 15 pixels. The adaptive was created by controlling the size of the elements based on the difference in the intensity of the pixels.