Difference between revisions of "Main Page"
Spyridon97 (talk | contribs) (→COVID-19) |
(→Announcements) |
||
| (97 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
| − | + | [[File:Thanksgiving lunch 2019.jpg|600px|thumb|frameless|center| Chrisochoides' Real-Time Computing (or in short CRTC) group before the pandemic.]] | |
| − | + | == Announcements == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ''' Professor Chrisochoides will be teaching (Fall’23) a new class on Quantum Computing; for more details, see [[https://crtc.cs.odu.edu/images/8/85/CS495-595_on_QC.png here.]] | |
| − | |||
| − | |||
| − | + | — Two students from this class got internships in Navy Research Lab and DoE’s Jefferson Lab''' | |
| − | + | === Congratulations to Dr. Thomadakis=== | |
| − | + | [[File:Polykarpos-Grad.jpeg|200px|left]] | |
| − | + | Congratulations to Dr. Thomadakis for his impressive Ph.D. work on parallel intelligent runtime software systems for HPC adaptive and irregular applications and Domain-Specific Languages. | |
| − | |||
| − | |||
| − | File: | ||
| − | |||
| − | |||
| − | + | The AI part of his PhD work ([https://crtc.cs.odu.edu/pub/html_output/details.php?&title=De-noising%20drift%20chambers%20in%20CLAS12%20using%20convolutional%20auto%20encoders&paper_type=2&from=year] [https://crtc.cs.odu.edu/pub/html_output/details.php?&title=Using%20Machine%20Learning%20for%20Particle%20Track%20Identification%20in%20the%20CLAS12%20Detector&paper_type=2&from=year] [https://crtc.cs.odu.edu/pub/html_output/details.php?&title=Track%20Identification%20for%20CLAS12%20using%20Artificial%20Intelligence&paper_type=1&from=year]) was completed in collaboration with [https://www.linkedin.com/in/gagik-gavalian-55926725/ Dr. Gagik Gavalian] from [https://www.jlab.org/ Jefferson Lab] and other former members of the [https://crtc.cs.odu.edu/Main_Page CRTC], including current member Kevin Garner. '''Some of the findings from his research is projected to have a significant impact over the next decade, with a value of approximately $100 million.''' | |
| − | |||
| − | |||
| − | + | Nuclear Physicists confirmed these results in a [https://www.odu.edu/article/ai-collaboration-between-odu-jefferson-lab-improves-data-analysis recent article], and Dr. Gavalian presented the findings in [https://indico.jlab.org/event/459/contributions/11745/attachments/9491/13757/CHEP-05-2023-Gavalian.pdf CHEP'23]. [https://www.odu.edu/directory/lawrence-weinstein Dr. Lawrence Weinstein], an ODU Eminent Scholar, professor of physics, and past chair of the Jefferson Lab Users Organization acknowledged in a recent article ([https://www.odu.edu/article/ai-collaboration-between-odu-jefferson-lab-improves-data-analysis]) that the project has improved particle trajectory reconstruction, resulting in a 35% increase in the number of reconstructed complicated nuclear collisions. This means that "more physics from the same data" is possible, which would have otherwise cost about $5 million per year. Dr. Gavalian's new results show that there has been an increase in statistics for physics observables from 35% to approximately 50-80% by adding the AI-based denoising ([https://crtc.cs.odu.edu/pub/html_output/details.php?&title=De-noising%20drift%20chambers%20in%20CLAS12%20using%20convolutional%20auto%20encoders&paper_type=2&from=year] ) of the data in the workflow, which adds another $5M per year to the earlier estimate by Dr. Weinstein. | |
| − | + | Next, Dr. Gagik will be looking into applying lessons learned to other Jefferson Lab Halls (sensors) and we will be putting this AI work on “steroids” using scalable D-NISQ Quantum Machine Learning. Our preliminary results ([https://crtc.cs.odu.edu/pub/html_output/details.php?&title=Evaluation%20of%20Scalable%20Quantum%20and%20Classical%20Machine%20Learning%20for%20Particle%20Tracking%20Classification%20in%20Nuclear%20Physics&paper_type=1&from=year] [https://crtc.cs.odu.edu/pub/html_output/details.php?&title=Scalable%20Quantum%20Edge%20Detection%20Method%20for%20D-NISQ%20Imaging%20Simulations:%20Use%20Cases%20from%20Nuclear%20Physics%20and%20Medical%20Image%20Computing&paper_type=1&from=year]) in Quantum Computing indicate that this is feasible. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | These accomplishments would not have been possible without the small investment from Jefferson Lab ($100K) and the Richard T. Cheng Endowment ($50K), as well as the help of many others, including the Dragas Foundation, which has assisted Olga Karadimou, an exceptional Ph.D. student at ODU, in staying at ODU with Polykarpos. | |
| − | |||
| − | |||
| − | === | + | === Protein-to-Protein Interactions === |
| − | + | Progress on protein-to-protein interactions using geometric feature analysis: [[COVID-19]]. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | + | A slideshow of Molecular Geometric Feature Analysis for SARS-CoV-2: click [[Molecular Geometric Feature Analysis | here]]. |
== Introduction == | == Introduction == | ||
| − | Finite Element (FE) mesh generation is a critical component for many (bio-)engineering and science applications. CRTCLab is developing a novel framework for highly scalable and energy-efficient high-quality mesh generation for the Finite Element Analysis in three and four dimensions. CRTC effort and focus is on research activities that combine domain-and application-specific knowledge with run-time system support to improve energy efficiency and scalability of parallel FE mesh generation codes. Traditionally, parallel FE mesh generation methods and software are developed without considering the architectural features of the supercomputer platforms on which they are eventually used for production. The main reason is the complexity of sequential, and moreover parallel, mesh generation algorithms. As a result, it is too expensive, in terms of labor and time, to customize the performance of parallel mesh generation software for specific supercomputing architectures. CRTC’s basic research is funded from NSF, NASA, NRA, total $2M and target (bio-)engineering applications are: [[#Extreme-Scale_Parallel_Mesh_Generation_&_Adaptation | Aerospace Industry Applications]] and [[#Medical_Image_Computing | Health Care Industry Applications]]. | + | Finite Element (FE) mesh generation is a critical component for many (bio-)engineering and science applications. CRTCLab is developing a novel framework for highly scalable and energy-efficient high-quality mesh generation for the Finite Element Analysis in three and four dimensions. CRTC effort and focus is on research activities that combine domain-and application-specific knowledge with run-time system support to improve energy efficiency and scalability of parallel FE mesh generation codes. Traditionally, parallel FE mesh generation methods and software are developed without considering the architectural features of the supercomputer platforms on which they are eventually used for production. The main reason is the complexity of sequential, and moreover parallel, mesh generation algorithms. As a result, it is too expensive, in terms of labor and time, to customize the performance of parallel mesh generation software for specific supercomputing architectures. CRTC’s basic research is funded from NSF, NASA, NRA, total $2M and target (bio-)engineering applications are: [[#Extreme-Scale_Parallel_Mesh_Generation_&_Adaptation | Aerospace Industry Applications]] and [[#Medical_Image_Computing | Health Care Industry Applications]]. [https://patents.google.com/?inventor=Nikos+CHRISOCHOIDES Six patent applications were filled.] |
<center> | <center> | ||
Latest revision as of 01:17, 26 January 2024
Contents
Announcements
Professor Chrisochoides will be teaching (Fall’23) a new class on Quantum Computing; for more details, see [here.]
— Two students from this class got internships in Navy Research Lab and DoE’s Jefferson Lab
Congratulations to Dr. Thomadakis
Congratulations to Dr. Thomadakis for his impressive Ph.D. work on parallel intelligent runtime software systems for HPC adaptive and irregular applications and Domain-Specific Languages.
The AI part of his PhD work ([2] [3] [4]) was completed in collaboration with Dr. Gagik Gavalian from Jefferson Lab and other former members of the CRTC, including current member Kevin Garner. Some of the findings from his research is projected to have a significant impact over the next decade, with a value of approximately $100 million.
Nuclear Physicists confirmed these results in a recent article, and Dr. Gavalian presented the findings in CHEP'23. Dr. Lawrence Weinstein, an ODU Eminent Scholar, professor of physics, and past chair of the Jefferson Lab Users Organization acknowledged in a recent article ([5]) that the project has improved particle trajectory reconstruction, resulting in a 35% increase in the number of reconstructed complicated nuclear collisions. This means that "more physics from the same data" is possible, which would have otherwise cost about $5 million per year. Dr. Gavalian's new results show that there has been an increase in statistics for physics observables from 35% to approximately 50-80% by adding the AI-based denoising ([6] ) of the data in the workflow, which adds another $5M per year to the earlier estimate by Dr. Weinstein.
Next, Dr. Gagik will be looking into applying lessons learned to other Jefferson Lab Halls (sensors) and we will be putting this AI work on “steroids” using scalable D-NISQ Quantum Machine Learning. Our preliminary results ([7] [8]) in Quantum Computing indicate that this is feasible.
These accomplishments would not have been possible without the small investment from Jefferson Lab ($100K) and the Richard T. Cheng Endowment ($50K), as well as the help of many others, including the Dragas Foundation, which has assisted Olga Karadimou, an exceptional Ph.D. student at ODU, in staying at ODU with Polykarpos.
Protein-to-Protein Interactions
Progress on protein-to-protein interactions using geometric feature analysis: COVID-19.
A slideshow of Molecular Geometric Feature Analysis for SARS-CoV-2: click here.
Introduction
Finite Element (FE) mesh generation is a critical component for many (bio-)engineering and science applications. CRTCLab is developing a novel framework for highly scalable and energy-efficient high-quality mesh generation for the Finite Element Analysis in three and four dimensions. CRTC effort and focus is on research activities that combine domain-and application-specific knowledge with run-time system support to improve energy efficiency and scalability of parallel FE mesh generation codes. Traditionally, parallel FE mesh generation methods and software are developed without considering the architectural features of the supercomputer platforms on which they are eventually used for production. The main reason is the complexity of sequential, and moreover parallel, mesh generation algorithms. As a result, it is too expensive, in terms of labor and time, to customize the performance of parallel mesh generation software for specific supercomputing architectures. CRTC’s basic research is funded from NSF, NASA, NRA, total $2M and target (bio-)engineering applications are: Aerospace Industry Applications and Health Care Industry Applications. Six patent applications were filled.
Principles for Building Practical and Scalable Domain-Specific Environment: Adaptive Data-Driven Mesh Generation for Modeling & Simulations
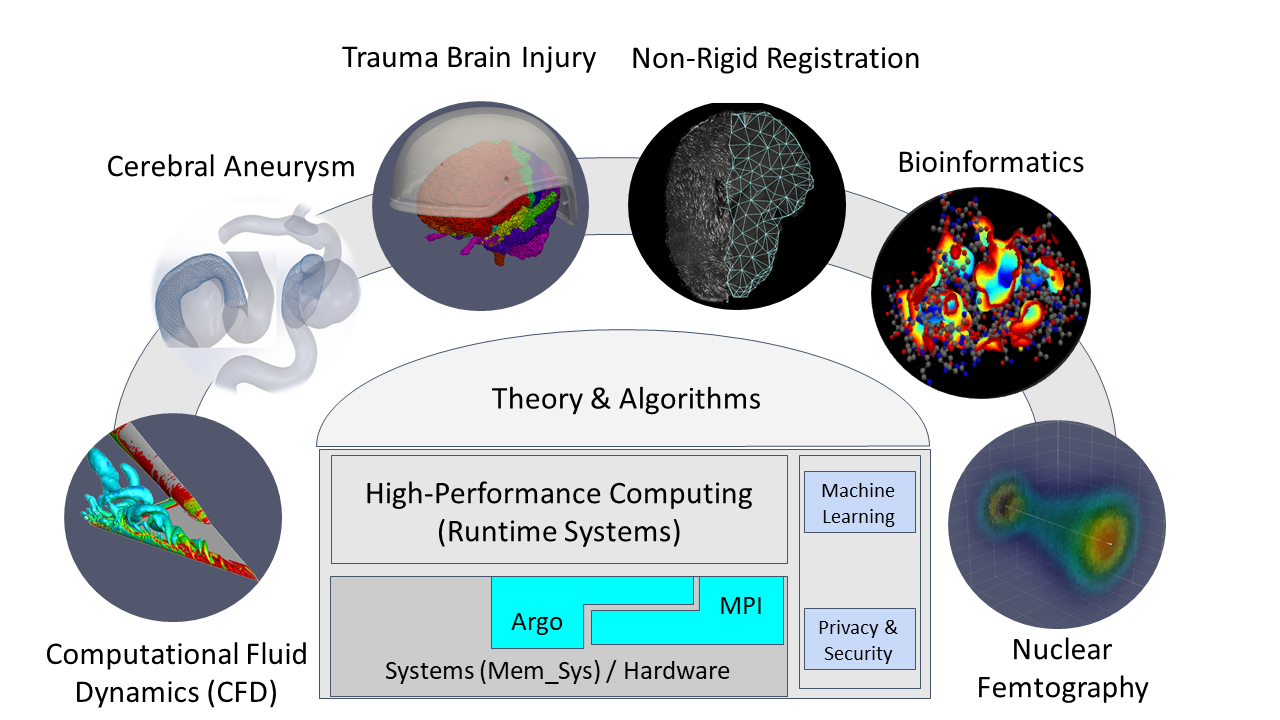
Broader Impact
The proposed activity will have a significant and broader impact on research and education, well beyond the bounds of this specific project and its external collaborators. Other than the direct impact to parallel unstructured mesh generation and runtime systems for supercomputers like Blue Waters, this project will benefit the performance evaluation community. This project will also promote the understanding on CerebroVascular Disease (CVD, or stroke), Image Guided Surgery for Caner, and DBS for Parkisons disease, all of which are leading natural death causes in the US.
Intellectual Merit The Intellectual Merit of this project is summarized in terms of three novel contributions: (1) an understanding of the complex computation, communication, and memory utilization patterns in a telescopic superposition of multiple parallel mesh generation codes, (2) new abstractions to efficiently throttle concurrency and to implement race-to-halt execution models at multiple levels of granularity, while orchestrating intra-and inter-layer memory management, data movement, and load balancing to improve energy-efficiency of memory and interconnect components, and (3) a novel approach to leveraging off-the-shelf and state-of-the-art low core meshing codes for highly parallel simulations.
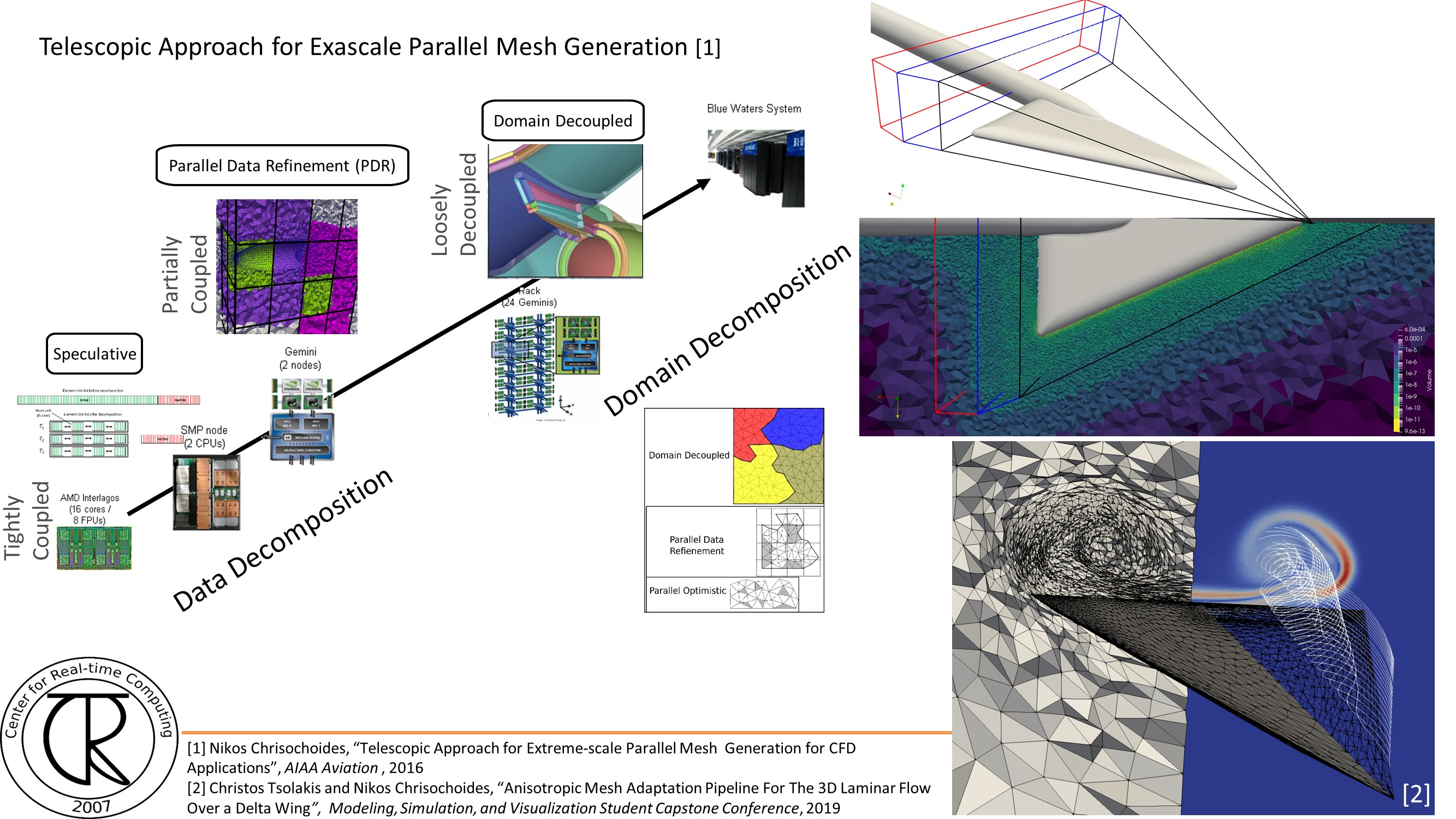
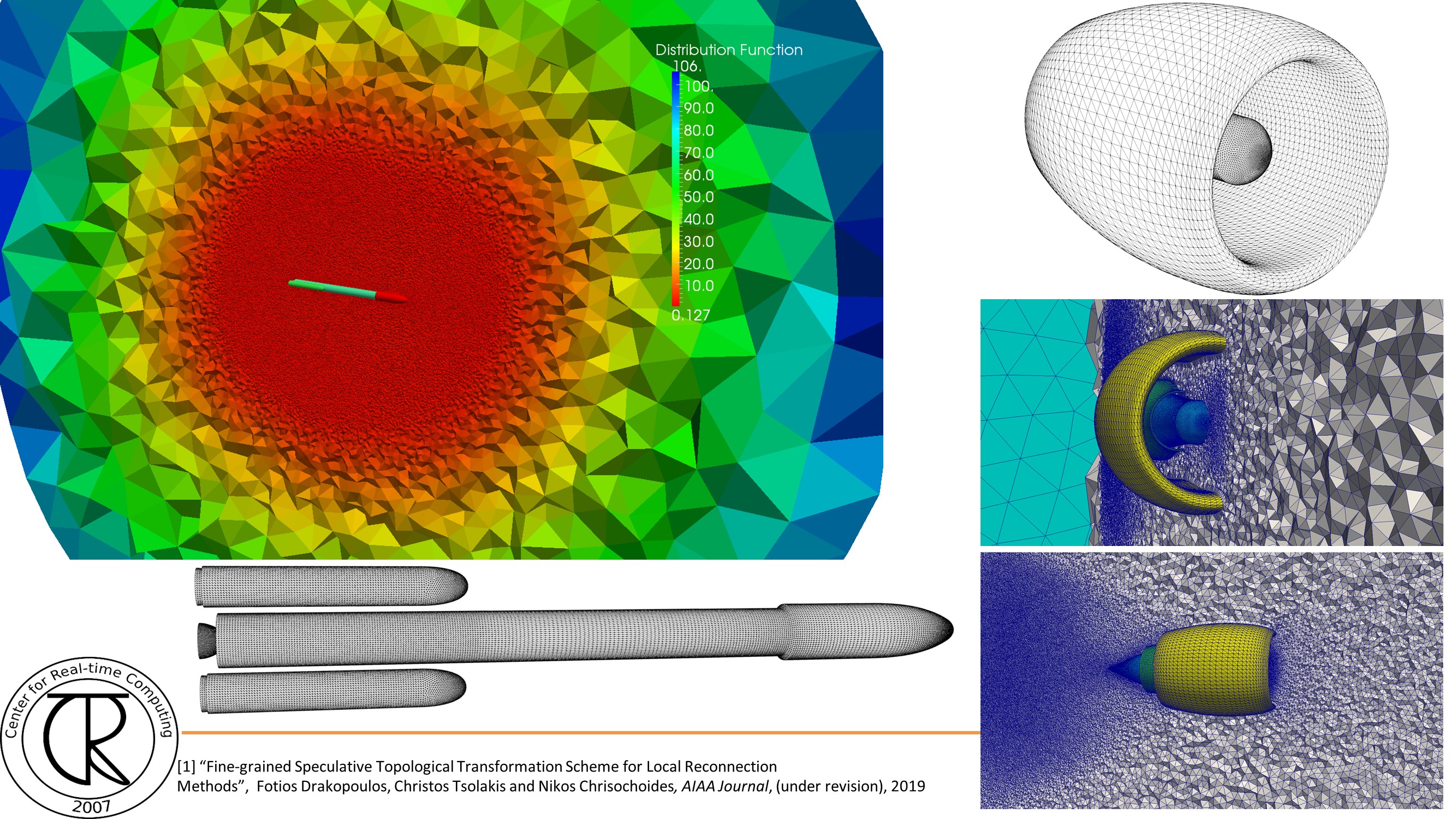
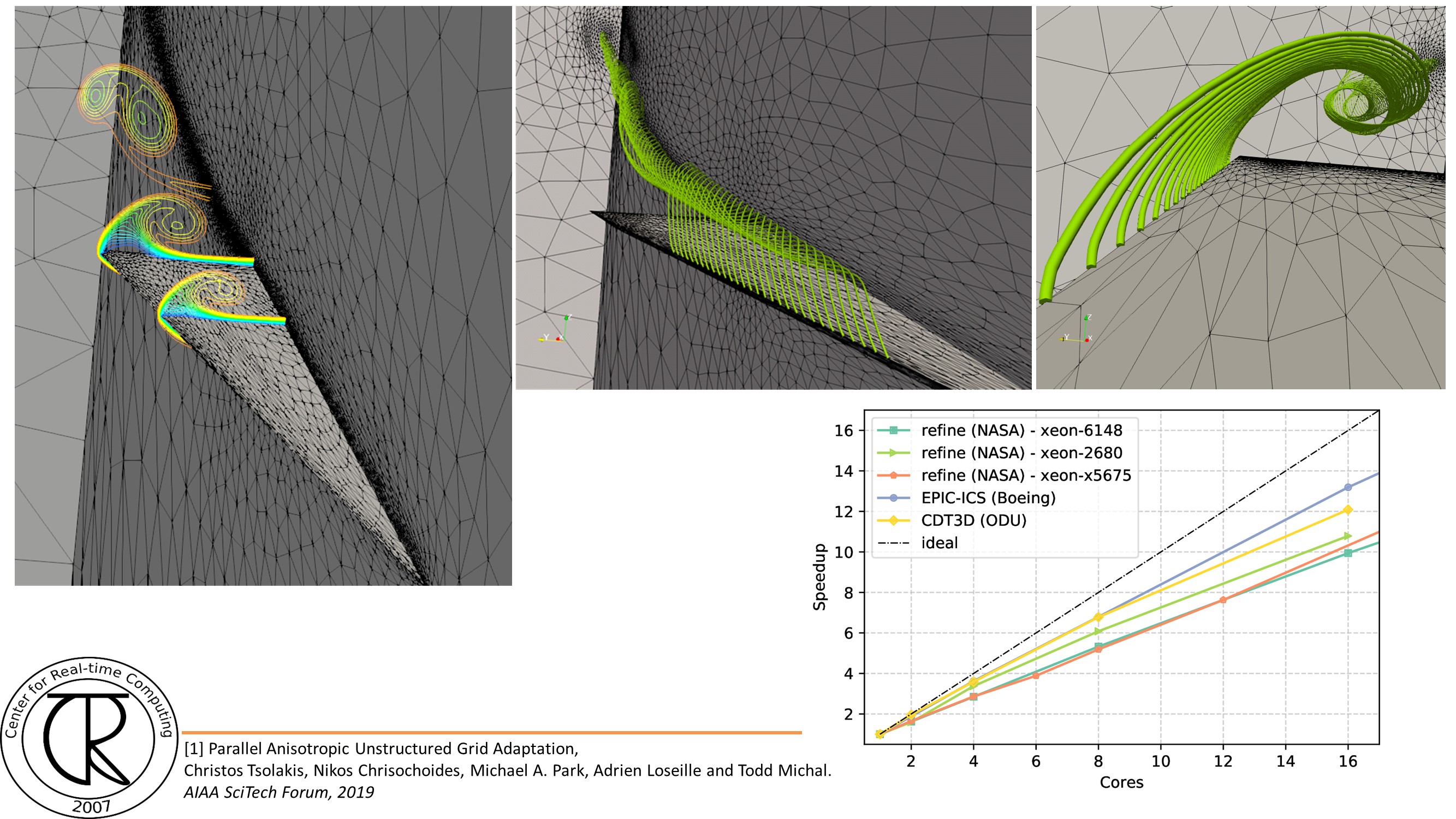
Extreme-Scale Adaptive/Anisotropic Parallel Mesh Generation
- CDT3D Meshes using speculative execution model at the chip level
Overview
Finite Element Mesh Generation is a critical component for many (bio-) engineering and science applications. The goal of this project is to deliver a novel Telescopic framework for highly scalable and energy efficient codes. Domain-and application-specific knowledge and run-time system support are combined to improve accuracy of FE computations.
CRTCLab addresses in depth two pillars: (i) high performance computing software runtime systems and (ii) mesh generation & adaptation based on error-metrics for NASA’s Computational Fluid Dynamics (CFD) Vision 2030. Specifically, CRTC’s focus is on the following three objectives: (1) design a multi-layered algorithmic and software framework for 3D tetrahedral anisotropic parallel mesh generation using state-of-the-art functionality supported by algorithms from AFLR and CRTC’s telescopic approach for parallel mesh generation , (2) develop error-based metrics adaptive anisotropic FE mesh generation methods and (3) design a power-aware parallel runtime software system for extreme-scale adaptive CFD computations including: (i) mesh generation & adaptation, and (ii) consistent error-based metrics for adaptation of any CFD discretizations with localizable error estimates. This is a joined project with NASA/LaRC and MIT.
We have assembled a team of established leaders (see External Collaborators) that are currently developing state-of-the-art work on mesh generation and adaptivity issues relevant to NASA’s CFD 2030 Vision and will broadly impact end-user productivity of users throughout DoD and NASA.
Objectives
- Design a multi-layered algorithmic and software framework for 3D tetrahedral anisotropic parallel mesh generation methods using state-of-the-art functionality supported by methods implemented in AFLR and CRTC’s telescopic approach for parallel mesh generation.
- Development of error-based metrics to drive an anisotropic adaptive process
- Design a power-aware parallel runtime software system for extreme-scale adaptive CFD computations including: (i) mesh generation & adaptation, and (ii) consistent error-based metrics for adaptation of any CFD discretization with localizable error estimates.
Real-time Adaptive Mesh Generation in Medical Image Computing
This application is using adaptive (isotropic) mesh refinement to improve (in real-time in the context of image-guided neurosurgery) the accuracy of 3D Physics-Based Deformable Registration of intra-operative MRI with pre-operative MRI.
Overview
- Develop adaptive deformable registration method for Deep Brain Stimulation (DBS). DBS is an effective palliative therapy for patients suffering from Essential Tremor, Parkinson’s disease, and other neurological movement disorders. Modern DBS surgery makes use of stereotactic systems and image guidance to accurately place electrode leads, as well as intra-operative imaging to surveil the location of the leads and guide the surgery. The effectiveness of DBS is directly correlated with the accuracy of DBS electrode lead placement, with more accurate electrode placement leading to better clinical outcomes. This project is a joined with VCU, VA.
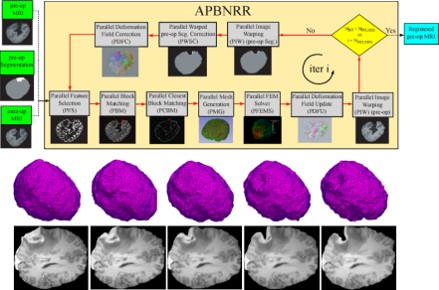
- Develop a new Adaptive Physics-Based Non-Rigid Registration (APBNRR) method developed for image guided brain tumor resection. Existing evaluation indicates that the the registration accuracy and performance of APBNRR are proved sufficient to be of clinical value in the operating room. Malignant gliomas are the most common primary and metastatic brain tumors. Treatment typically includes surgical removal followed by radiotherapy or chemotherapy. Total resection is difficult to achieve during tumor removal because of the infiltrative nature of gliomas and because brain tumors are often embedded in critical functional brain tissue. In image-guided neurosurgery, co-registered pre-operative anatomical, functional, and diffusion tensor imaging can be used to facilitate a maximally safe resection of brain tumors in certain areas of the brain. However, because the brain can deform significantly during surgery, particularly in the presence of tumor resection, non-rigid registration of the preoperative image data to the patient is required. This is a joined project with Harvard Medical School, MA.
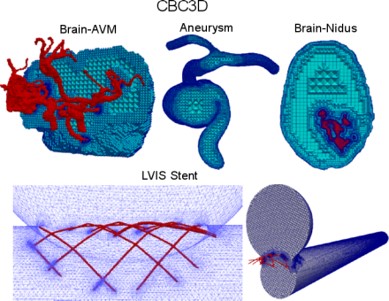
- Test five new available (in the clinic) neurovascular devices that have not been compared before through angiographic washout analysis and accurate micro CT-based tessellation (i.e., mesh or grid reconstruction) and CFD analysis in idealized sidewall aneurysm models. The CFD simulations and angiographic washout analysis will be performed on flow diverter implantations in actual patient geometries. There is a need for robust and easy-to-use Image-to-Mesh (I2M) conversion methods and software that streamline the discretization of such complex vascular geometries with fine structures with the CFD modeling process. In this project we will focus on the development and validation of fully functional I2M conversion algorithms and software (i.e., tools) for complex vascular geometries with fine structures like stents. This is a joined project with Medical School at Stony Brook University, NY.